- 1Laboratorio de Citogenética de Peixes, Departamento de Genética e Evolução, Universidade Federal de São Carlos (UFSCar), São Carlos, Brazil
- 2Museu de Zoologia da Universidade de São Paulo (MZUSP), São Paulo, Brazil
- 3Laboratório de Sistemática e Morfologia de Peixes, Departamento de Sistemática e Ecologia (DSE), Universidade Federal da Paraíba (UFPB), João Pessoa, Brazil
- 4Laboratório de Gentética Animal, Instituto Nacional de Pesquisa da Amazônia, Coordenação de Biodiversidade, Manaus, Brazil
- 5Laboratório de Biologia Cromossômica, Estrutura e Função, Departamento de Biologia Estrutural, Molecular e Genética, Universidade Estadual de Ponta Grossa, Ponta Grossa, Brazil
- 6Institute of Human Genetics, University Hospital Jena, Jena, Germany
Miniature fishes have always been a challenge for cytogenetic studies due to the difficulty in obtaining chromosomal preparations, making them virtually unexplored. An example of this scenario relies on members of the family Lebiasinidae which include miniature to medium-sized, poorly known species, until very recently. The present study is part of undergoing major cytogenetic advances seeking to elucidate the evolutionary history of lebiasinids. Aiming to examine the karyotype diversification more deeply in Pyrrhulina, here we combined classical and molecular cytogenetic analyses, including Giemsa staining, C-banding, repetitive DNA mapping, comparative genomic hybridization (CGH), and whole chromosome painting (WCP) to perform the first analyses in five Pyrrhulina species (Pyrrhulina aff. marilynae, Pyrrhulina sp., P. obermulleri, P. marilynae and Pyrrhulina cf. laeta). The diploid number (2n) ranged from 40 to 42 chromosomes among all analyzed species, but P. marilynae is strikingly differentiated by having 2n = 32 chromosomes and a karyotype composed of large meta/submetacentric chromosomes, whose plesiomorphic status is discussed. The distribution of microsatellites does not markedly differ among species, but the number and position of the rDNA sites underwent significant changes among them. Interspecific comparative genome hybridization (CGH) found a moderate divergence in the repetitive DNA content among the species’ genomes. Noteworthy, the WCP reinforced our previous hypothesis on the origin of the X1X2Y multiple sex chromosome system in P. semifasciata. In summary, our data suggest that the karyotype differentiation in Pyrrhulina has been driven by major structural rearrangements, accompanied by high dynamics of repetitive DNAs.
Introduction
Characiformes comprise a very diverse and abundant freshwater order (Nelson et al., 2016), in which the family Lebiasinidae is represented by 75 valid species (Fricke et al., 2021) widely distributed across South and Central America (Weitzman and Weitzman, 2003). The phylogenetic relationships of the Lebiasinidae remained in doubt for a long time, but more recent phylogenetic analysis indicate their proximity to the Ctenoluciidae (Calcagnotto et al., 2005; Oliveira et al., 2011), which was also reinforced by the different studies (Arcila et al., 2017; Betancur-R et al., 2019; Melo et al., 2021). Most Lebiasinidae species reach about 60 mm of Standard Length (SL), but miniature species, not surpassing a maximum of 26 mm SL, is found within the Pyrrhulininae, whereas medium-sized species up to 150 mm SL can be found within Lebiasininae (Weitzman and Weitzman, 2003).
Because of their small sizes and difficulties in obtaining good chromosomal preparations, species of Lebiasinidae were, for a long time, little analyzed in terms of cytogenetics, with scarce references mainly on the chromosomal number of few species (Scheel, 1973; Oliveira et al., 1991; Arai, 2011). However, this scenario has recently undergone significant changes with the methodological advance of cytogenetics and its applicability among small to miniature fishes of Pyrrhulina, Lebiasina, Copeina, and Nannostomus genus (de Moraes et al., 2017, de Moraes et al.,2019; Sassi et al., 2019; Toma et al., 2019; Sassi et al., 2020; Sember et al., 2020).
Pyrrhulina is one of the most speciose genera of the subfamily Pyrrhulininae, with 19 valid small species (Fricke et al., 2021), ranging from 30.4 to 85 mm SL (Weitzman and Weitzman, 2003; Netto-Ferreira and Marinho, 2013). The genus is among the most problematic, with many poorly known species, species complexes, and old taxonomic problems (Netto-Ferreira and Marinho, 2013). The first Pyrrhulina species to have some chromosomal data evidenced was Pyrrhulina cf. australis, with 2n = 40 chromosomes, mainly acrocentric ones (Oliveira et al., 1991). Taxonomic boundaries of P. australis are still poorly defined, demonstrated in subsequent studies (de Moraes et al., 2017; de Moraes et al.,2019) of two morphotypes. Both P. australis and Pyrrhulina aff. australis showed similar data 2n = 40 (4st + 36a), distinct from P. brevis, 2n = 42 (2sm + 4st + 36a), with no evidence of heteromorphic sex chromosomes in the three species (de Moraes et al., 2017; de Moraes et al., 2019). Another species, P. semifasciata, was analyzed, presenting 2n = 42 (4st + 38a) in females, and 2n = 41 (1m + 4st + 36a) in males, the latter with three unpaired chromosomes because of a multiple X1X1X2X2/X1X2Y sex chromosome system (de Moraes et al., 2019). This occurrence was also confirmed by comparative genomic hybridizations (CGH) and whole-chromosome painting (WCP), with some indications that the Y chromosome originated by centric fusions of non-homologous acrocentric chromosomes (de Moraes et al., 2019).
To improve the knowledge of the evolutionary processes within the genus Pyrrhulina, we combined classical and molecular cytogenetic analyses, including Giemsa staining, C-banding, repetitive DNA mapping, comparative genomic hybridization (CGH), and whole chromosome painting (WCP to perform the first analyses in five Pyrrhulina species (Pyrrhulina aff. marilynae, Pyrrhulina sp., P. obermulleri, P. marilynae and Pyrrhulina cf. laeta). The results highlighted relationships and particular evolutionary paths at the chromosomal and genomic levels among the species. In addition, the hypothesis on the origin of the multiple sex chromosome system in P. semifasciata is validated.
Materials and Methods
Animals
The collection sites, number, and sex of the specimens investigated are presented in Figure 1, Table 1. Part of the sampling (Figure 1, white circles) resembles the one previously analyzed by de Moraes et al. (2017), de Moraes et al. (2019) with different cytogenetic and molecular methods. Animals were collected with the authorization of the Brazilian environmental agency ICMBIO/SISBIO (license no. 48628-14) and SISGEN (A96FF09). All species were properly identified by morphological criteria, and the specimens were deposited in the fish collection of the Museu de Zoologia da Universidade de São Paulo (MZUSP) under the voucher numbers (119077, 119079, 123073, 123080) and the Universidade Federal da Paraíba (UFPB) museum under the voucher number (12079, 12080, 12082 and 12083). Experiments followed ethical and anesthesia conducts and were approved by the Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos (process number CEUA 1853260315).
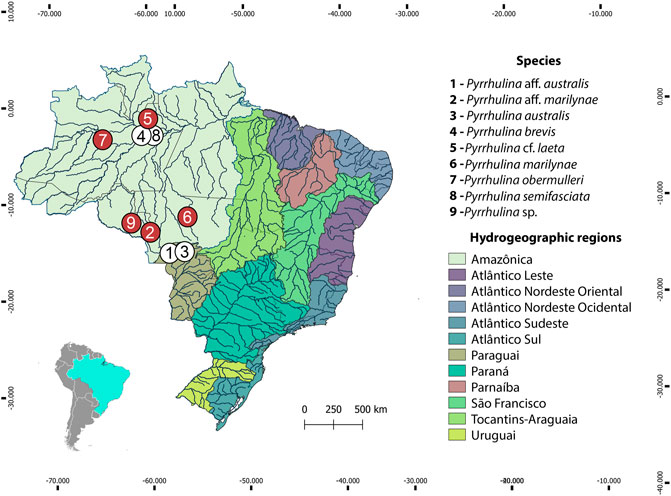
FIGURE 1. Brazilian collection sites of the Pyrrhulina species cytogenetically investigated in the present study (red circles) and the ones previously cytogenetically analyzed (white circles: data from (de Moraes et al., 2017; de Moraes et al., 2019).

TABLE 1. Geographical coordinates, sample size, and diploid number of Pyrrhulina (Characiformes, Lebiasinidae) species collected in Brazil.
Chromosomal Preparations and Analysis of the Constitutive Heterochromatin
Mitotic chromosomes were obtained from kidney cells by the protocol described in Bertollo et al. (2015). The distribution of constitutive heterochromatin was observed by the C-banding methodaccording to (Sumner, 1972).
Repetitive DNA Mapping with Fluorescence in situ Hybridization (FISH)
The 5S rDNA probe included 120 base pairs (bp) of the 5S rDNA gene coding region and 200 bp of non-transcribed spacer (NTS) (Pendás et al., 1994). The 18S rDNA probe was composed of a 1,400-bp-long segment of the 18S rDNA coding region (Cioffi et al., 2009). Both probes were directly labeled with the Nick-Translation Mix Kit (Jena Bioscience, Jena, Germany)—18S rDNA with ATTO488-dUTP and 5S rDNA with ATTO550-dUTP, according to the manufacturer’s instructions. The (CA)15, (GA)15, (CGG)10 microsatellite probes were directly labeled with Cy3 during the synthesis, according to Kubat et al. (2008). In addition, since it contains the lowest 2n, telomeric (TTAGGG)n sequence was also used as probe in P. marylinae. This probe was generated by PCR in the absence of a template according to Ijdo et al. (1991) and later labeled with ATTO550-dUTP with the Nick-Translation Mix Kit (Jena Bioscience, Jena, Germany). FISH experiments followed the methodology described in Yano et al. (2017). Metaphase chromosomes were treated with RNAse A (40 μg/ml) for 1.5 h at 37°C and the DNA denatured in 70% formamide/2× SSC at 72°C for 3.15 min. A hybridization mixture (2.5 ng/μL probes, 50% deionized formamide, 10% dextran sulfate) was then dropped on the slides, and the hybridization process was performed overnight at 37°C in a moist chamber. The first post-hybridization wash was performed with 1× SSC for 5 min at 65°C, followed by the second one performed with 4xSSC/Tween for 5 min, at room temperature. Chromosomes were then counterstained with DAPI, and the slides were mounted with an antifade solution (Vectashield from Vector Laboratories, Burlingame, CA).
FISH for Whole Chromosome Painting
As P. semifasciata represents the only Pyrrhulina species that harbors an X1X2Y multiple sex system, a Y-chromosome probe, named PSEMI-Y, was previously prepared by microdissection, as described in (de Moraes et al., 2019) Male and female metaphases of P. marilynae, Pyrrhulina aff. marilynae, Pyrrhulina sp., P. obermulleri, Pyrrhulina cf. laeta were used for Zoo-FISH experiments with the PSEMI-Y probe, according to procedures described in Yano et al. (2017). The hybridization was performed for 72 h at 37°C in a moist chamber, with post-hybridization washes with 1xSSC for 5 min at 65°C, and in 4xSSC/Tween (RT). 10 μg of male-derived C0t-1 DNA from P. semifasciata was used as suppressor in each experiment. Chromosomes were stained with DAPI (1.2 μg/ml) and the slides were mounted with an antifade solution, as described above.
Probes for Comparative Genomic Hybridization
The genomic DNAs (gDNAs) from male and female specimens of P. marilynae, Pyrrhulina aff. marilynae, Pyrrhulina sp., P. obermulleri, Pyrrhulina cf. laeta, P. australis, Pyrrhulina aff. australis, P. brevis, and P. semifasciata were extracted from liver tissue by the standard phenol-chloroform-isoamyl alcohol method (Sambrook and Russell, 2001). For intraspecific comparisons, the male-derived gDNAs of all species were labeled with ATTO550-dUTP and the female gDNAs with ATTO 488-dUTP, by nick translation (Jena Bioscience, Jena, Germany). The repetitive sequences were blocked using unlabeled C0t-1 DNA in all experiments, according to (Zwick et al., 1997). The final hybridization mixture for each slide (20 μL) was composed of male- and female-derived gDNAs (500 ng each), plus 25 μg of female-derived C0t-1 DNA from the respective species. The probe was ethanol-precipitated, and the dry pellets were mixed in a hybridization mixture containing 50% formamide, 2× SSC, 10% SDS, 10% dextran sulfate, and Denhardt´s buffer, pH 7.0.
For interspecific comparisons, the gDNA of male specimens of P. australis (Paus), Pyrrhulina aff. australis (Pafa), P. semifasciata (Psem), P. brevis (Pbre), P. marilynae (Pmar), Pyrrhulina aff. marilynae (Pafm), Pyrrhulina sp. (Psp), P.obermulleri (Pobe) and Pyrrhulina cf. laeta (Pcfl) were hybridized against metaphase chromosomes of P. marilynae. This species was selected since it harbors the lowest 2n = 32 until now register for the genus, coupled with a remarkable karyotype differentiation. For this purpose, male-derived gDNA of P. marilynae was labeled with ATTO 550-dUTP, while the gDNAs of the other species were labeled with ATTO 488-dUTP (P. australis, Pyrrhulina aff. marilynae, P. brevis and P. obermulleri) or ATTO 425-dUTP (Pyrrhulina aff. australis, Pyrrhulina sp., P. semifasciata and Pyrrhulina cf. laeta), both through nick translation (Jena Bioscience, Jena, Germany).
The interspecific comparisons were divided into a set of four slides. In the first slide, the final probe mixture was composed of 500 ng of male-derived gDNA plus 10 μg of male-derived C0t-1 DNA of each of the following species: P. marilynae, P. australis, and Pyrrhulina aff. australis. In the second slide, the final probe mixture was composed of 500 ng of male-derived gDNA plus 10 μg of male-derived C0t-1 DNA of each one of the following species: P. marilynae, Pyrrhulina aff. marilynae and Pyrrhulina sp. In the third slide, the final probe mixture was composed of 500 ng of male-derived gDNA plus 10 μg of male-derived C0t-1 DNA of each one of the following species: P. marilynae, P. brevis, and P. semifasciata. Finally, in the fourth slide, the final probe mixture was composed of 500 ng of male-derived gDNA plus and 10 μg of male-derived C0t-1 DNA of each one of the following species: P. marilynae, P. obermulleri, and Pyrrhulina cf. laeta. The chosen ratio of probe vs. C0t-1 DNA amount was based on previous experiments performed in our fish studies (de Moraes et al., 2019; Toma et al., 2019; Sassi et al., 2020). The CGH experiments followed the methodology described in Sember et al. (2018).
Microscopy and Images Processing
To confirm the diploid number, karyotype structure and FISH results inat least 30 metaphase spreads were analyzed per individual. The microscopy images were captured using an Olympus BX50 epifluorescence microscope (Olympus Corporation, Ishikawa, Japan) coupled with a CoolSNAP camera, and the images were processed using Image-Pro Plus 4.1 Software (Media Cybernetics, Silver Spring, MD, United States). Final images were optimized and arranged using Adobe Photoshop, version CC 2020. Chromosomes were classified as metacentric (m), submetacentric (sm), subtelocentric (st), or acrocentric (a), according to their arm ratios (Levan, 1964). As the males and females results showed no differences, only male metaphases were represented in the figures.
Results
Karyotypes and Heterochromatin Distribution
The diploid number ranged from 2n = 40 to 42 among the following four species: Pyrrhulina sp. (2n = 40; 2st+38a), Pyrrhulina aff. marilynae (2 = 40; 40a), P. obermulleri (2n = 42; 2m/sm+8st+32a) and Pyrrhulina cf. laeta (2n = 42; 2m/sm+4st+36a), the two latter also sharing a characteristic small metacentric/submetacentric pair. On the other hand, P. marilynae differed by presenting a very distinct karyotype composition (2n = 32; 8m/sm+4st+20a). These results represent the first cytogenetic data for the abovementioned species. The constitutive heterochromatin was distributed at the pericentromeric region of several chromosome pairs in P. marilynae and Pyrrhulina aff. marilynae. In its turn, Pyrrhulina sp., P. obermulleri, and Pyrrhulina cf. laeta presented a remarkable series of interstitial and pericentromeric C-bands, in addition to telomeric ones (Figure 2). In our sampling, we did not observe any karyotype differences between males and females.

FIGURE 2. Male and female karyotypes of Pyrrhulina marilynae(A, F, and K), Pyrrhulina aff. marilynae(B, G and L), Pyrrhulina sp. (C, H and M), P. obermulleri(D, I and N) and Pyrrhulina cf. laeta(E, J and O) arranged after Giemsa staining (A-E), C-banding (F-J), and dual-color in situ hybridization (FISH) with 18S (green) and 5S (red) ribosomal DNA probes (K-O). Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI).Scale bar = 5 μm.
Chromosomal Mapping of Repetitive DNA Sequences
All the five species differ by the distribution of the multigene rDNA families. Pyrrhulina sp. and P. marilynae were the only species with only one chromosome pair bearing 18S rDNA sites, found at the telomeric region of acrocentric pairs 4 and 9, respectively. Six to twelve centromeric or telomeric sites occur in the other three species, including bitelomeric sites in Pyrrhulina aff. marilynae (pair 11) and Pyrrhulina cf. laeta (pairs 6 and 13). As for the 5S rDNA, from six to twelve centromeric sites occured among species, including a syntenic condition for the 5S and 18S rDNA repeats in the chromosome pair 6 of Pyrrhulina cf. laeta, the same pair that displays bitelomeric 18S rDNA signals in this species (Figure 2). The distribution of the microsatellites (CA)15, (GA)15, and (CGG)10 does not differ significantly among species, having a preferential location in the centromeric and telomeric regions of the chromosomes, in addition to some interstitial sites. However, (CA)15 differs quantitatively, with a greater number of conspicuous sites compared to the other microsatellites, especially in Pyrrhulina aff. marilynae and Pyrrhulina cf. laeta. In the same way, (CGG)10 occurs in smaller amounts in the five species (Figure 3). The (TTAGGG)n repeats showed the expected hybridization signals on telomeres of P. marylinae (Figure 4F). Whole chromosome painting–WCP.
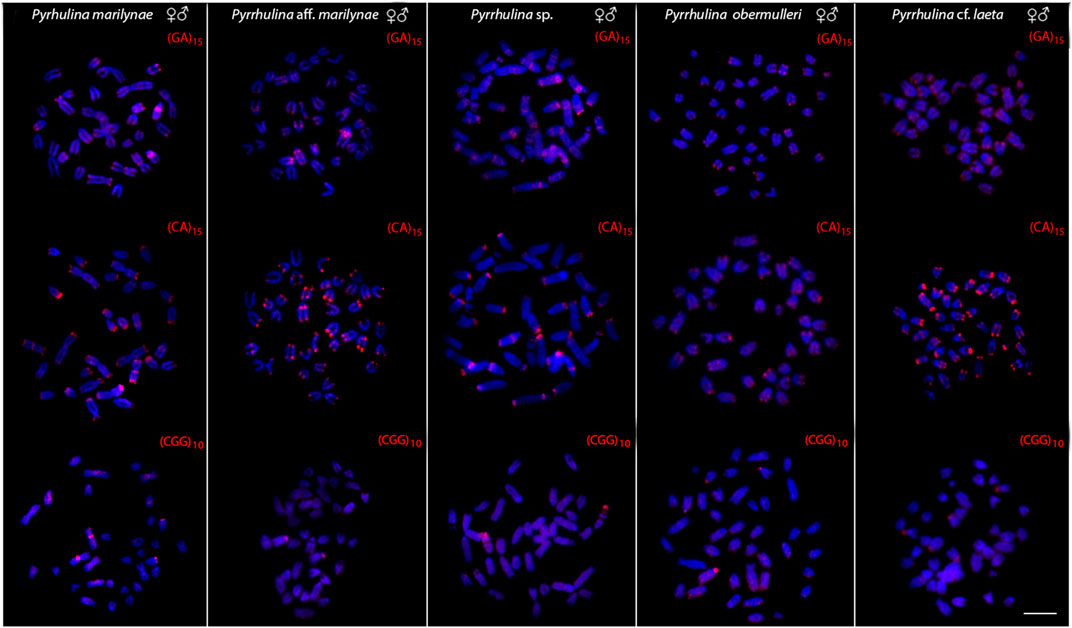
FIGURE 3. Male and female metaphase plates of Pyrrhulina marilynae; Pyrrhulina aff. marilynae; Pyrrhulina sp.; P. obermulleri and Pyrrhulina cf. laeta shows the general distribution of the microsatellites (GA)15, (CA)15 and (CGG)10 on chromosomes. Bar = 5 μm.
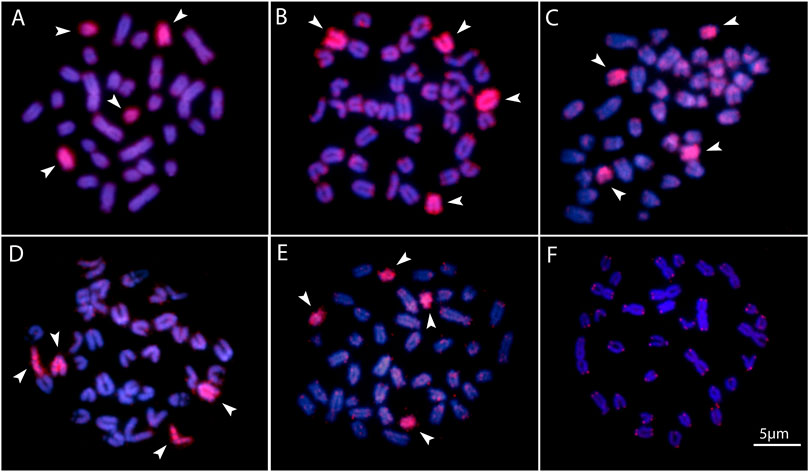
FIGURE 4. Zoo-FISH with the PSEMI-Y probeon male metaphase plates of P. marilynae(A), Pyrrhulina aff. marilynae (B), Pyrrhulina cf. laeta(C), Pyrrhulina sp. (D), and P. obermulleri(F) shows the distribution of the telomeric (TTAGGG)n repeats in P. marilynae. Bar = 5 μm.
Two acrocentric chromosome pairs were entirely painted with the PSEMI-Y probe in Pyrrhulina marilynae, P. obermulleri, Pyrrhulina sp., Pyrrhulina aff. marilynae and Pyrrhulina cf. laeta (Figures 4A–E).
Comparative Genomic Hybridization–CGH
The interespecific genomic comparison among Pyrrhulina marilynae and other Pyrrhulina species (P. semifasciata, P. australis, P. brevis, P. obermulleri, Pyrrhulina aff. australis, Pyrrhulina sp., Pyrrhulina aff. marilynae, Pyrrhulina cf. laeta) revealed a high level of DNA compartmentalization, within all species presenting a distinct composition of repetitive DNA sequences and specific signals. However, P. marilynae shows more evident species-specific arrangements when compared to the other species. (Figure 5). Intraspecific genomic hybridization between males and females did not show any clustering for sex-specific sequences in all species (data not shown).
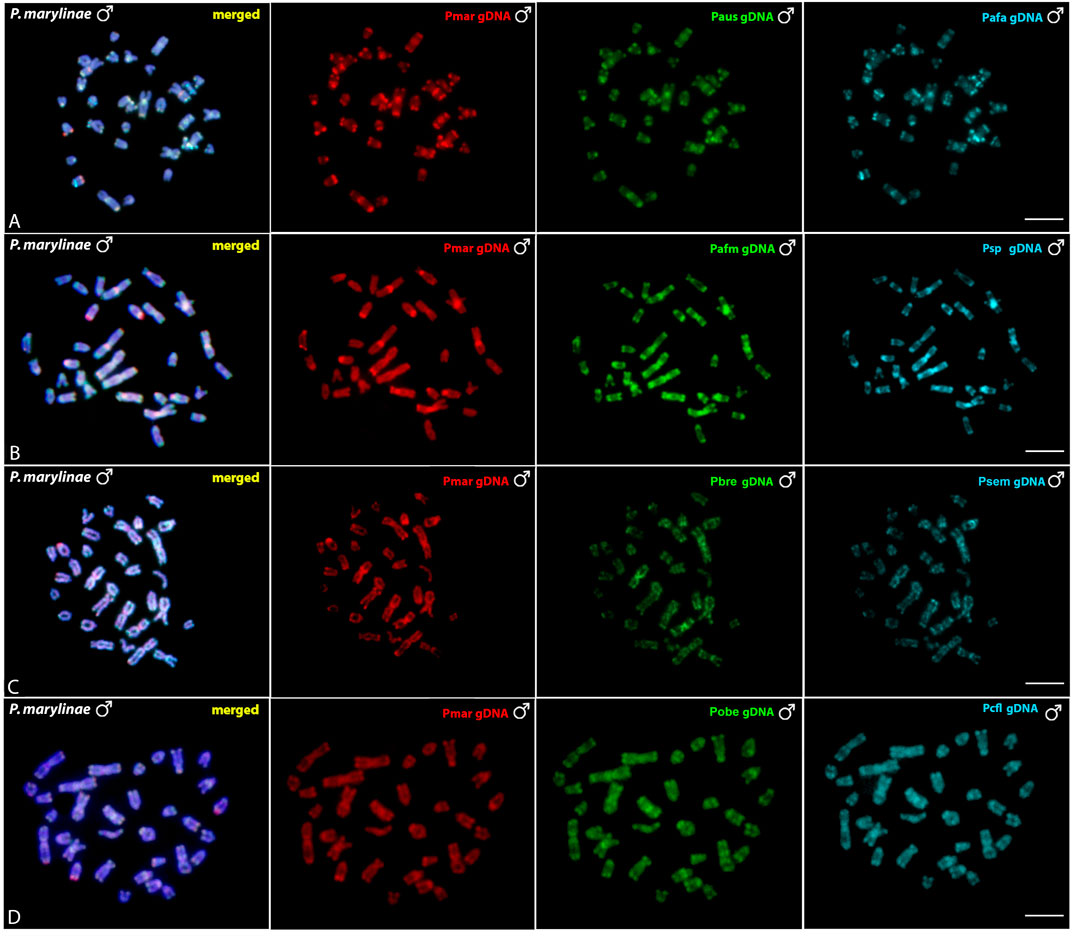
FIGURE 5. Comparative genomic hybridization (CGH) using male-derived genomic probes from Pyrrhulina species hybridized onto male chromosomes of P. marilynae. The common genomic regions are depicted in the 1st column in each line representing the experiments A-D. Hybridization between the gDNA of P. marilynae (Pmar), P. australis (Paus) and Pyrrhulina aff. australis (Pafa) (A); P. marilynae(Pmar), Pyrrhulina aff. marilynae (Pafm) and Pyrrhulina sp. (Psp) (B); P. marilynae(Pmar), P. brevis (Pbre) and P. semifasciata (Psem) (C); P. marilynae (Pmar), P. obermulleri (Pobe) and Pyrrhulina cf. laeta (Pcfl) (D). Bar = 5 μm.
Discussion
Overall, two main evolutionary trends are proposed for the karyotypic evolution of the Lebiasinidae: 1) the conservation of a plesiomorphic karyotype in the subfamily Lebiasininae, with 2n = 36 bi-armed chromosomes and, 2) high variations in diploid numbers and karyotypic structures in the subfamily Pyrrhulininae, with the predominance of acrocentric chromosomes (Sassi et al., 2020). It is noteworthy that the karyotypic structure of Lebiasininae, 2n = 36 biarmed chromosomes, is similar to that found in the sister family Ctenoluciidae (de Souza e Sousa et al., 2017; Sassi et al., 2019; de Souza e Sousa et al., 2021). Therefore, in this scenario, the majority of the acrocentric chromosomes found in the species of the Pyrrhulininae are probably derived from rearrangements such as centric fissions (Sassi et al., 2020). However, unlike other Pyrrhulina species, P. marilynae has the smallest 2n identified in the genus so far, 2n = 32, including four typical meta/submetacentric pairs. Some exceptions within the subfamily showed secondary fusion events of acrocentric chromosomes giving rise to metacentric chromosomes, reducing the diploid number as observed in Nannostomus unifasciatus (Sember et al., 2020). Biarmed chromosomes could also represent remnants of the ancestral karyotype condition that were maintained during the evolutionary processes. However, no ITS was found in any chromosome of P. marilynae, but such a scenario does not exclude the hypothesis of fusion, given that telomeric regions can be lost after the rearrangement occurs (Bolzán, 2017). Thus, to corroborate such hypotheses and to determine whether the evolutionary trajectory of karyotype change in Pyrrhulina is directed mainly towards centric fusions or fissions, cytogenetic data should be discussed in a larger phylogenetic framework of interspecific and intergeneric relationships of Lebiasinidae.
CGH procedures have greatly assisted cytogenetic studies (Symonová et al., 2013; Cioffi et al., 2017; Cioffi et al.,2019), as among all Pyrrhulina studied so far. In fact, despite showing close genomic similarities, the species also show considerable divergences, in addition to species-specific repetitive DNA and C-band patterns, thus helping to understand their differential evolutionary paths, considering the taxonomic problems still pending in this fish group. In addition, multiple and syntenic ribosomal sites are not frequently observed among fishes, but these chromosomal features are very informative cytotaxonomic markers regarding Pyrrhulininae species. Comparatively, they occur more frequently among Pyrrhulina than in other species of this subfamily (de Moraes et al., 2017; de Moraes et al.,2019; Sassi et al., 2019; Sassi et al.,2020; Toma et al., 2019; Sember et al., 2020). Like Pyrrhulina aff. australis (de Moraes et al., 2017), Pyrrhulina sp., and P. marilynae present multiple 5S rDNA sites and only one 18S rDNA site, thus differentiating them from Pyrrhulina aff. marilynae, P. obermulleri, and Pyrrhulina cf. laeta, as well as from some other Pyrrhulina species (de Moraes et al., 2017; de Moraes et al.,2019), which have multiple 5S and 18S rDNA sites. Furthermore, the syntenic condition for the 18S/5S rDNAs in Pyrrhulina cf. laeta is shared with P. brevis and P. australis, indicating a high rDNA diversity. (Figure 6). In its turn, the 18S rDNA clusters are distributed on distal chromosome positions for all investigated Pyrrhulina species (de Moraes et al., 2017; de Moraes et al.,2019; this study), as also occur among Copeina (Toma et al., 2019), Lebiasina (Sassi et al., 2019), and Nannostomus (Sember et al., 2020), so as in the species of the sister family, Ctenoluciidae (de Souza e Sousa et al., 2017; de Souza e Sousa et al., 2021).

FIGURE 6. Representative idiograms of Pyrrhulina species showing the distribution of the 18S (green) and 5S rDNA (red) sites on chromosomes, based on the present study and some other previous data (de Moraes et al., 2017; de Moraes et al., 2019). Bar = 5 μm.
Microsatellite distribution patterns have significantly contributed to evolutionary studies in fish species, especially regarding sex chromosome differentiation (Kubat et al., 2008; Cioffi et al., 2012; Terencio et al., 2012; Kejnovský et al., 2013; Poltronieri et al., 2014; Yano et al., 2014; de Freitas et al., 2018). Among the five Pyrrhulina species now investigated, as well as in other previous analyzed ones (de Moraes et al., 2017; de Moraes et al.,2019), the distribution of the microsatellites did not significantly differ among them, although the (CA)15 repeats present a greater number of more conspicuous sites than the other microsatellites, especially in Pyrrhulina aff. marilynae and Pyrrhulina cf. laeta. It is noteworthy that microsatellites have a preferential location in the telomeric and centromeric regions of fish chromosomes (Cioffi and Bertollo, 2012), as occur with the (CA)15 and (GA)15 motifs in Pyrrhulina, despite some interstitial and pericentromeric signs in Pyrrhulina cf. laeta, P. marilynae, Pyrrhulina aff. marilynae and Pyrrhulina sp., thus differentiating these species from others previously studied (de Moraes et al., 2017; de Moraes et al.,2019). Furthermore, it is also frequent that microsatellites and other repetitive sequences occur in the association among fish (Cioffi and Bertollo, 2012), such as in Hepsetus odoe (Carvalho et al., 2017), Lebiasina bimaculata (Sassi et al., 2019), and Silurichthys phaiosoma (Ditcharoen et al., 2020), for example. This is the scenario that also occurs in Pyrrhulina sp., in which the (CGG)10 microsatellite located in the telomeric region of pair 4 shares the same chromosomal region with 18S rDNA repeats.
Fish, besides presenting high diversity in morphological and genetic characteristics, also have a variety of sex chromosome systems (Sember et al., 2021). About nine differentiated systems, involving the XX/XY and ZZ/ZW sex chromosomes and their variations, have been identified among species, including several Neotropical ones (Sember et al., 2021). It is noteworthy that among the multiple systems, the ♀X1X1X2X2/♂X1X2Y is the most prevalent one, and commonly originated by centric or tandem fusions of the ancestral Y with an autosomal member of the karyotype, giving rise to neo-Y chromosomes, as identified in a variety of fish species (Sember et al., 2021). That includes P. semifasciata, the only Lebiasinidae representative highlighting heteromorphic sex chromosomes so far (de Moraes et al., 2019), in addition to a putative ZZ/ZW sex system present in Lebiasina bimaculata (Sassi et al., 2019). Although our intraspecific CGH results in the current analyzed species did not reveal any sex-specific differentiated region, our WCP experiment with the Y-derived probe of P. semifasciata entirely painted two acrocentric pairs, suggesting that putative proto-XY chromosomes may occur in these species. Thus, it supports our previous hypothesis on the origin of the P. semifasciata sex chromosome system through centric fusion between the non-homologous acrocentric, giving rise to the large metacentric Y chromosome. That can be considered as an apomorphy of this species when compared to others of the genus. Furthermore, the experiments also showed that although the karyotype of P. marilynae has large metacentric chromosomes, these do not correspond to the heteromorphic sex chromosome of P. semifasciata (Figure 4).
Conclusion
Our data advances the understanding of evolutionary trends of the Lebiasinidae, particularly concerning Pyrrhulina. Karyotypes with 2n = 40–42, with the predominance of mono-armed chromosomes, are more frequent among its species, except for P. marilynae, which has a smaller diploid number (2n = 32), and several atypical biarmed chromosomes, a characteristic that differentiates this species from the others analyzed in the genus. However, we cannot rule out the hypothesis that this karyotypic reduction (2n = 32) may have been generated by secondary fusions that allowed the formation of the four meta/submetacentric pairs identified in P. marilynae. The present data also highlighted the putative proto-XY chromosomes that may occur in these species and support the occurrence, through centric fusion, of the multiple sex chromosome system of P. semifasciata as an independent evolutionary event of this Lebiasinidae species. Our results highlight the importance of chromosomal data as valuable markers for understanding the evolutionary relationships among Lebiasinidae species.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos (process number CEUA 1853260315).
Author Contributions
RM and MC carried out the cytogenetic analysis and drafted the manuscript. TH, AA-R, and PV helped in the cytogenetics analysis, drafted and revised the manuscript. TL, GD, FS, VO, EF and MM drafted and revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
MC was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (Proc. No. 302449/2018-3) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (Proc. No. 2020/11772-8). RM and FS were supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (Proc. No. 2019/25045-3 and 2020/02681-9, respectively). This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Finance Code 001.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful for FAPESP and CNPq. We also appreciate the contributions in the collection of animals from Hugmar Pains, Alany Pedrosa Gonçalves (Mamirauá Sustainable Development Reserve) and Ezequiel Aguiar de Oliveira.
References
Arai, R. (2011). Fish Karyotypes: A Check List. Berlin, Germany: Springer Science & Business Media. doi:10.1007/978-4-431-53877-6
Arcila, D., Ortí, G., Vari, R., Armbruster, J. W., Stiassny, M. L. J., Ko, K. D., et al. (2017). Genome-wide Interrogation Advances Resolution of Recalcitrant Groups in the Tree of Life. Nat. Ecol. Evol. 1, 1–10. doi:10.1038/s41559-016-0020
Cioffi, M. B., Molina, W. F., Artoni, R. F., and Bertollo, L. A. C. (2012). Chromosomes as Tools for Discovering Biodiversity — The Case of Erythrinidae Fish Family,” in Recent Trends in Cytogenetic Studies—Methodologies and Applications. Editor P. Tirunilai (Rijeka: IntechOpen), 125–146. doi:10.5772/35890
Bertollo, L. A. C., Cioffi, M., and Moreira-Filho, O. (2015). “Direct Chromosome Preparation from Freshwater Teleost Fishes,” in Fish Cytogenet. Tech. (Chondrichthyans Teleosts). Editors C. Ozouf-Costaz, E. Pisano, F. For, and L. F Almeida Toledo, 31–36. doi:10.1201/b18534-6
Betancur‐R., R., Arcila, D., Vari, R. P., Hughes, L. C., Oliveira, C., Sabaj, M. H., et al. (2019). Phylogenomic Incongruence, Hypothesis Testing, and Taxonomic Sampling: The Monophyly of Characiform Fishes. Evolution 73, 329–345. doi:10.1111/evo.13649
Bolzán, A. D. (2017). Interstitial Telomeric Sequences in Vertebrate Chromosomes: Origin, Function, Instability and Evolution. Mutat. Research/Reviews Mutat. Res. 773, 51–65. doi:10.1016/j.mrrev.2017.04.002
Calcagnotto, D., Schaefer, S. A., and DeSalle, R. (2005). Relationships Among Characiform Fishes Inferred from Analysis of Nuclear and Mitochondrial Gene Sequences. Mol. Phylogenet. Evol. 36, 135–153. doi:10.1016/j.ympev.2005.01.004
Carvalho, P. C., de Oliveira, E. A., Bertollo, L. A. C., Yano, C. F., Oliveira, C., Decru, E., et al. (2017). First Chromosomal Analysis in Hepsetidae (Actinopterygii, Characiformes): Insights into Relationship between African and Neotropical Fish Groups. Front. Genet. 8, 203. doi:10.3389/fgene.2017.00203
Cioffi, M. B., and Bertollo, L. A. C. (2012). Chromosomal Distribution and Evolution of Repetitive DNAs in Fish. Repetitive DNA 7, 197–221. doi:10.1159/000337950
Cioffi, M. B., Martins, C., Centofante, L., Jacobina, U., and Bertollo, L. A. C. (2009). Chromosomal Variability Among Allopatric Populations of Erythrinidae Fish Hoplias malabaricus: Mapping of Three Classes of Repetitive DNAs. Cytogenet. Genome Res. 125, 132–141. doi:10.1159/000227838
Cioffi, M. d. B., Yano, C. F., Sember, A., and Bertollo, L. A. C. (2017). Chromosomal Evolution in Lower Vertebrates: Sex Chromosomes in Neotropical Fishes. Genes 8, 258. doi:10.3390/genes8100258
Cioffi, M. de. B., Ráb, P., Ezaz, T., Bertollo, L., Lavoué, S., Oliveira, E., et al. (2019). Deciphering the Evolutionary History of Arowana Fishes (Teleostei, Osteoglossiformes, Osteoglossidae): Insight from Comparative Cytogenomics. Int J Mol Sci 20, 4296. doi:10.3390/ijms20174296
de Freitas, N. L., Al-Rikabi, A. B. H., Bertollo, L. A. C., Ezaz, T., Yano, C. F., de Oliveira, E. A., et al. (2018). Early Stages of XY Sex Chromosomes Differentiation in the Fish Hoplias malabaricus (Characiformes, Erythrinidae) Revealed by DNA Repeats Accumulation. Curr Genomics. 19, 216–226. doi:10.2174/1389202918666170711160528
de Moraes, R. L. R., Bertollo, L. A. C., Marinho, M. M. F., Yano, C. F., Hatanaka, T., Barby, F. F., et al. (2017). Evolutionary Relationships and Cytotaxonomy Considerations in the GenusPyrrhulina(Characiformes, Lebiasinidae). Zebrafish 14, 536–546. doi:10.1089/zeb.2017.1465
de Moraes, R. L. R., Sember, A., Bertollo, L. A. C., De Oliveira, E. A., Ráb, P., Hatanaka, T., et al. (2019). Comparative Cytogenetics and Neo-Y Formation in Small-Sized Fish Species of the Genus Pyrrhulina (Characiformes, Lebiasinidae). Front. Genet. 10, 678. doi:10.3389/fgene.2019.00678
de Souza e Sousa, J. F., Viana, P. F., Bertollo, L. A. C., Cioffi, M. B., and Feldberg, E. (2017). Evolutionary Relationships Among Boulengerella Species (Ctenoluciidae, Characiformes): Genomic Organization of Repetitive DNAs and Highly Conserved Karyotypes. Cytogenet. Genome Res. 152, 194–203. doi:10.1159/000480141
Ditcharoen, S., Sassi, F. d. M. C., Bertollo, L. A. C., Molina, W. F., Liehr, T., Saenjundaeng, P., et al. (2020). Comparative Chromosomal Mapping of Microsatellite Repeats Reveals Divergent Patterns of Accumulation in 12 Siluridae (Teleostei: Siluriformes) Species. Genet. Mol. Biol. 43. doi:10.1590/1678-4685-GMB-2020-0091
Ijdo, J. W., Wells, R. A., Baldini, A., and Reeders, S. T. (1991). Improved Telomere Detection Using a Telomere Repeat Probe (TTAGGG)ngenerated by PCR. Nucl. Acids Res. 19, 4780. doi:10.1093/nar/19.17.4780
Kejnovský, E., Michalovova, M., Steflova, P., Kejnovska, I., Manzano, S., Hobza, R., et al. (2013). Expansion of Microsatellites on Evolutionary Young Y Chromosome. PLoS One 8, e45519. doi:10.1371/journal.pone.0045519
Kubat, Z., Hobza, R., Vyskot, B., and Kejnovsky, E. (2008). Microsatellite Accumulation on the Y Chromosome inSilene Latifolia. Genome 51, 350–356. doi:10.1139/G08-024
Levan, A., Fredga, K., and Sandberg, A. A. (1964). Nomenclature for Centromeric Position on Chromosomes. Hereditas 52, 201–220. doi:10.1111/j.1601-5223.1964.tb01953.x
Melo, B. F., Sidlauskas, B. L., Near, T. J., Roxo, F. F., Ghezelayagh, A., Ochoa, L. E., et al. (2021). Accelerated Diversification Explains the Exceptional Species Richness of Tropical Characoid Fishes. Syst. Biol., 1–21. doi:10.1093/sysbio/syab040
Nelson, J. S., Grande, T. C., and Wilson, M. V. H. (2016). Fishes of the World. Hoboken, NJ: John Wiley & Sons. doi:10.2307/2412830
Netto-Ferreira, A. L., and Marinho, M. M. F. (2013). New Species of Pyrrhulina (Ostariophysi: Characiformes: Lebiasinidae) from the Brazilian Shield, with Comments on a Putative Monophyletic Group of Species in the Genus. Zootaxa 3664, 369–376. doi:10.11646/zootaxa.3664.3.7
Oliveira, C., Andreata, A. A., Toledo, L. F. A., and Toledo, S. A. (1991). Karyotype and Nucleolus Organizer Regions of Pyrrhulina Cf Australis (Pisces, Characiformes, Lebiasinidae). Rev. Bras. Genética, 14, 685–690.
Oliveira, C., Avelino, G. S., Abe, K. T., Mariguela, T. C., Benine, R. C., Ortí, G., et al. (2011). Phylogenetic Relationships within the Speciose Family Characidae (Teleostei: Ostariophysi: Characiformes) Based on Multilocus Analysis and Extensive Ingroup Sampling. BMC Evol. Biol. 11, 1–25. doi:10.1186/1471-2148-11-275
Pendás, A. M., Moran, P., Freije, J. P., and Garcia-Vazquez, E. (1994). Chromosomal Mapping and Nucleotide Sequence of Two Tandem Repeats of Atlantic salmon 5S rDNA. Cytogenet. Cel Genet 67, 31–36. doi:10.1159/000133792
Poltronieri, J., Marquioni, V., Bertollo, L. A. C., Kejnovsky, E., Molina, W. F., Liehr, T., et al. (2014). Comparative Chromosomal Mapping of Microsatellites inLeporinusSpecies (Characiformes, Anostomidae): Unequal Accumulation on the W Chromosomes. Cytogenet. Genome Res. 142, 40–45. doi:10.1159/000355908
R. Fricke, W. N. Eschmeyer, and R. van der Laan (Editors) (2021). Eschmeyer’s Catalog of Fishes: Genera, Species, References. Available at: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (Accessed September 2, 2021).
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning-Sambrook & Russel-Vol. 1, 2, 3. Cold Spring Harbor, NY: Cold Springs Harb. Lab. Press.
Sassi, F. de. M. C., Oliveira, E., Bertollo, L., Nirchio, M., Hatanaka, T., Marinho, M., et al. (2019). Chromosomal Evolution and Evolutionary Relationships of Lebiasina Species (Characiformes, Lebiasinidae). Int J Mol Sci 20, 2944. doi:10.3390/ijms20122944
Sassi, F. d. M. C., Hatanaka, T., Moraes, R. L. R. d., Toma, G. A., Oliveira, E. A. d., Liehr, T., et al. (2020). An Insight into the Chromosomal Evolution of Lebiasinidae (Teleostei, Characiformes). Genes 11, 365. doi:10.3390/genes11040365
Sember, A., Bertollo, L. A. C., Ráb, P., Yano, C. F., Hatanaka, T., de Oliveira, E. A., et al. (2018). Sex Chromosome Evolution and Genomic Divergence in the Fish Hoplias malabaricus (Characiformes, Erythrinidae). Front. Genet. 9, 71. doi:10.3389/fgene.2018.00071
Sember, A., de Oliveira, E. A., Ráb, P., Bertollo, L. A. C., de Freitas, N. L., Viana, P. F., et al. (2020). Centric Fusions behind the Karyotype Evolution of Neotropical Nannostomus Pencilfishes (Characiforme, Lebiasinidae): First Insights from a Molecular Cytogenetic Perspective. Genes 11, 91. doi:10.3390/genes11010091
Sember, A., Nguyen, P., Perez, M. F., Altmanová, M., Ráb, P., and Cioffi, M. d. B. (2021). Multiple Sex Chromosomes in Teleost Fishes from a Cytogenetic Perspective: State of the Art and Future Challenges. Phil. Trans. R. Soc. B 376, 20200098. doi:10.1098/rstb.2020.0098
Souza, J., Guimarães, E., Pinheiro-Figliuolo, V., Cioffi, M. B., Bertollo, L. A. C., and Feldberg, E. (2021). Chromosomal Analysis of Ctenolucius Hujeta Valenciennes, 1850 (Characiformes): A New Piece in the Chromosomal Evolution of the Ctenoluciidae. Cytogenet. Genome Res. 161, 195–202. doi:10.1159/000515456
Sumner, A. T. (1972). A Simple Technique for Demonstrating Centromeric Heterochromatin. Exp. Cel Res. 75, 304–306. doi:10.1016/0014-4827(72)90558-7
Symonová, R., Majtánová, Z., Sember, A., Staaks, G. B., Bohlen, J., Freyhof, J., et al. (2013). Genome Differentiation in a Species Pair of Coregonine Fishes: an Extremely Rapid Speciation Driven by Stress-Activated Retrotransposons Mediating Extensive Ribosomal DNA Multiplications. BMC Evol. Biol. 13, 42–11. doi:10.1186/1471-2148-13-42
Terencio, M. L., Schneider, C. H., Gross, M. C., Nogaroto, V., de Almeida, M. C., Artoni, R. F., et al. (2012). Repetitive Sequences Associated with Differentiation of W Chromosome in Semaprochilodus Taeniurus. Genetica 140, 505–512. doi:10.1007/s10709-013-9699-4
Toma, G. A., de Moraes, R. L. R., Sassi, F. d. M. C., Bertollo, L. A. C., de Oliveira, E. A., Rab, P., et al. (2019). Cytogenetics of the Small-Sized Fish, Copeina Guttata (Characiformes, Lebiasinidae): Novel Insights into the Karyotype Differentiation of the Family. PLoS One 14, e0226746. doi:10.1371/journal.pone.0226746
Weitzman, M., and Weitzman, S. H. (2003). Family Lebiasinidae. Check List Freshw. fishes South Cent. Am. Porto Alegre, Edipucrs 729p, 241–250.
Yano, C. F., Bertollo, L. A. C., and Cioffi, M. D. B. (2017). “Fish-FISH: Molecular Cytogenetics in Fish Species,” in Fluorescence In Situ Hybridization (FISH). Berlin, Germany: Springer, 429–443. doi:10.1007/978-3-662-52959-1_44
Yano, C. F., Bertollo, L. A. C., Molina, W. F., Liehr, T., and de Bello Cioffi, M. (2014). Genomic Organization of Repetitive DNAs and its Implications for Male Karyotype and the Neo-Y Chromosome Differentiation in Erythrinus Erythrinus (Characiformes, Erythrinidae). Comp Cytogenet. 8, 139–151. doi:10.3897/compcytogen.v8i2.7597
Keywords: fishes, repetitive DNAs, karyotype evolution, sex chromosomes, evolution
Citation: de Moraes RLR, Sassi FdMC, Bertollo LAC, Marinho MMF, Viana PF, Feldberg E, Oliveira VCS, Deon GA, Al-Rikabi ABH, Liehr T and Cioffi MdB (2021) Tracking the Evolutionary Trends Among Small-Size Fishes of the Genus Pyrrhulina (Characiforme, Lebiasinidae): New Insights From a Molecular Cytogenetic Perspective. Front. Genet. 12:769984. doi: 10.3389/fgene.2021.769984
Received: 03 September 2021; Accepted: 22 September 2021;
Published: 06 October 2021.
Edited by:
Ricardo Utsunomia, Federal Rural University of Rio de Janeiro, BrazilReviewed by:
František Marec, Biology Centre of the Czech Academy of Sciences (CAS), Institute of Entomology, CzechiaDaniel Garcia-Souto, University of Vigo, Spain
Camilla Di Nizo, Zoological Research Museum Alexander Koenig (LG), Germany
Copyright © 2021 de Moraes, Sassi, Bertollo, Marinho, Viana, Feldberg, Oliveira, Deon, Al-Rikabi, Liehr and Cioffi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Liehr, Thomas.Liehr@med.uni-jena.de
 Renata Luiza Rosa de Moraes
Renata Luiza Rosa de Moraes Francisco de Menezes Cavalcante Sassi
Francisco de Menezes Cavalcante Sassi Luiz Antonio Carlos Bertollo
Luiz Antonio Carlos Bertollo Manoela Maria Ferreira Marinho
Manoela Maria Ferreira Marinho Patrik Ferreira Viana
Patrik Ferreira Viana Eliana Feldberg
Eliana Feldberg Vanessa Cristina Sales Oliveira
Vanessa Cristina Sales Oliveira Geize Aparecida Deon1,5
Geize Aparecida Deon1,5 Thomas Liehr
Thomas Liehr Marcelo de Bello Cioffi
Marcelo de Bello Cioffi