Population genetics of zig-zag eel (Mastacembelus armatus) uncover gene flow between an isolated island and the mainland China
- 1Key Laboratory of Prevention and Control for Aquatic Invasive Alien Species, Ministry of Agriculture and Rural Affairs, Guangdong Modern Recreational Fisheries Engineering Technology Center, Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 2Key Laboratory of Aquatic Animal Immune Technology of Guangdong Province, Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 3Department of Fisheries, Agro-Tech Extension Center of Guangdong Province, Guangzhou, China
Introduction: Mastacembelus armatus is a commercially valuable fish, normally distributed in southern China and Southeast Asia. The natural population size of M. armatus is shrinking in recent years because of overfishing and habitat loss. In order to clarify the genetic diversity and differentiation of M. armatus populations, we collected 114 samples from eight populations in southern China and Vietnam and analyzed their population structure using nuclear ribosomal DNA sequences, the concatenated 18S and ITS2 regions.
Methods: Genomic DNA from the fin clip was extracted and sequenced on an Illumina novaseq 6000 (Illumina, USA) high-throughput sequencing platform in accordance with the manufacturer’s instructions. After assembly and annotation, haplotype diversity, TCS network analysis, AMOVA analysis, population pairwise genetic distances, and UPGMA tree construction were conducted based on the concatenated sequences of 18S and ITS2.
Results and discussion: In total, eleven nrDNA haplotypes were detected based on the concatenated sequences of 18S and ITS2. Amongst, three haplotypes were the main haplotypes, as representatives of three corresponding Clusters. There were two major Clusters in China, however, the Cluster in Vietnam was significantly divergent from the other two in China, likely due to the lack of river connection between China and Vietnam. Interestingly, based on low FST value, we found that gene flow occurred between the isolated island, Hainan Province, and the mainland China of Guangxi Province, probably as a result of exposed continental shelf connected them during glacial periods. In general, combing our data and literature data, genetic diversity and differentiation of M. armatus populations are relatively high regardless of spatial scale, although their natural population size is declining. This suggests that it is not too late to adopt measures to protect M. armatus, which benefits not only species itself but also the whole ecosystem.
Introduction
Mastacembelus armatus, the common name is zig-zag eel or tire-track spiny eel, is an economically important fish, belonging to the Order Symbranchiformes (Family: Mastacembelidae; Genus: Mastacembelus). Among the four species of the genus, M. armatus is the largest (Serajuddin and Pathak, 2012). It is widely distributed in southern China, mainly in Yangtze River and Pearl River (Xue et al., 2020), and Southeast Asia, such as India, Thailand, Nepal, Vietnam, Sri Lanka, and Pakistan (Hossain et al., 2015; Gupta and Banerjee, 2016; Han et al., 2019). Usually, it inhabits rivers, streams, ponds, beels and inundated fields (Hossain et al., 2015; Gupta and Banerjee, 2016). M. armatus is a carnivorous fish, it prefers to feed on crustacean and insect larvae when young while the adults devour small fish and tadpoles (Hossain et al., 2015). M. armatus is in high demand on the market as it attracts consumers with its delicious taste, no intermuscular spines and high nutritional value (Gupta and Banerjee, 2016; Li et al., 2016; Xue et al., 2020). Besides, the appealing color pattern of M. armatus makes it a popular aquarium fish as well (Gupta and Banerjee, 2016).
However, due to overfishing and habitat loss, the wild population size of M. armatus gradually declines year by year (Hossain et al., 2012; Rahman et al., 2016; Xue, 2018). M. armatus is designated as an endangered species in Bangladesh (IUCN Bangladesh, 2000) and has been classified as least concern by the International Union for Conservation of Nature (IUCN) (IUCN, 2019). In addition, large-scale artificial breeding has not been achieved for M. armatus (Jiang, 2018). Therefore, it is urgently needed to clarify the present condition of natural populations of M. armatus, particularly their genetic diversity and structure, thus providing basis for their biological conservation. As a native species in China, M. armatus is assigned as a key protected wild aquatic animal by Fujian, Guangdong, and Hunan provinces, and moreover, a national germplasm resource reserve of M. armatus has been established in Fujian Province (Jiang, 2018). Furthermore, aquaculture of M. armatus has intensified in several provinces of China, greatly facilitating its artificial breeding (Han et al., 2017; Han et al., 2019).
So far, a lot of studies have been conducted on M. armatus, such as studies at individual level on their morphology (Shu et al., 2017; Zhou et al., 2019), nutritional composition (Wu et al., 2010; Fan et al., 2018), metal bioaccumulation (Javed and Usmani, 2016; Pandey et al., 2017), karyotype (Oliveira et al., 1997);, taxonomy (Jiang, 2018; Duong et al., 2020), reproduction (Serajuddin and Pathak, 2012), histopathology (Dhole et al., 2011), sex-specific markers (Xue et al., 2020; Xue et al., 2021b), artificial breeding (Lin et al., 2016; Xue, 2018), Toll-Like Receptors (TLR) (Han et al., 2017; Han et al., 2019), mitochondrial genome sequencing (Li et al., 2016; Han et al., 2018), whole genome sequences at the chromosomal scale (Xue et al., 2021a), as well as population studies (Wang et al., 2012; Zou, 2013; Chen, 2014; Yang et al., 2016; Lin, 2017; Jiang, 2018; Thapliyal et al., 2020; Gao et al., 2022). Among the above listed population studies, some used mitochondrial DNA markers, such as COI (Chen, 2014; Jiang, 2018; Gao et al., 2022), Cytb (Wang et al., 2012; Thapliyal et al., 2020; Gao et al., 2022) and D-loop (Chen, 2014), and others employed nuclear makers, like SSR (Zou, 2013; Yang et al., 2016; Lin, 2017) and exon-primed intron-crossing markers (EPICs) (Jiang, 2018). These studies, however, are all from Chinese populations, except one study from India (Thapliyal et al., 2020). Population data from Southeast Asia is very scarce. Therefore, we expanded our sampling area from China to Vietnam to fill our knowledge gap. Additionally, mitochondrial markers were mostly used in M. armatus population studies, only few nuclear markers like SSR and EPICs, while no reports were based on nuclear ribosomal DNA (nrDNA) markers to reveal their genetic diversity (Wang et al., 2012; Zou, 2013; Chen, 2014; Yang et al., 2016; Lin, 2017; Jiang, 2018; Gao et al., 2022). It is well known that nrDNA markers have been successfully employed in various fish population research (Mladineo et al., 2013; Garcia et al., 2015; Ağdamar and Tarkan, 2019), such as 18S, ITS1, ITS2 and 28S, due to their high evolutionary rates (Presa et al., 2002). Therefore, in this study, we employed the concatenated 18S and ITS2 sequences to assess levels of genetic diversity and differentiation of M. armatus populations from China and Vietnam at different spatial scale.
Materials and methods
Sampling
M. armatus from seven regions in southern China and one region in Vietnam were sampled in 2021. Sample sets collected from a single region were considered a population. In China, we collected M. armatus from four Provinces. Two populations were sampled from Guangdong Province, three populations from Guangxi Province, one population from Jiangxi Province and one population from Hainan Province (Table 1, Figure 1). What should be noted is that Hainan Province is located in an independent island, spatially separated with the mainland China by Qiongzhou Strait. Additionally, Guangxi Province is geographically adjacent with Vietnam. The Vietnam samples were collected from Guangzhou Lanhai Marine Technology Co., Ltd, Guangzhou city, Guangdong Province, China (latitude: 23.21°N, longitude: 113.47°E). More specifically, we collected 12 and 16 samples in GDHY and GDQY regions from Guangdong Province, respectively; 18, 18, 15 samples in GXBS, GXLZ and GXYL regions from Guangxi Province, respectively; 7 samples in HNHK region from Hainan Province, 13 samples in JXGZ region from Jiangxi Province, 15 samples in YN region from Vietnam. A total of 114 samples from eight populations were collected in this study (Table 1).
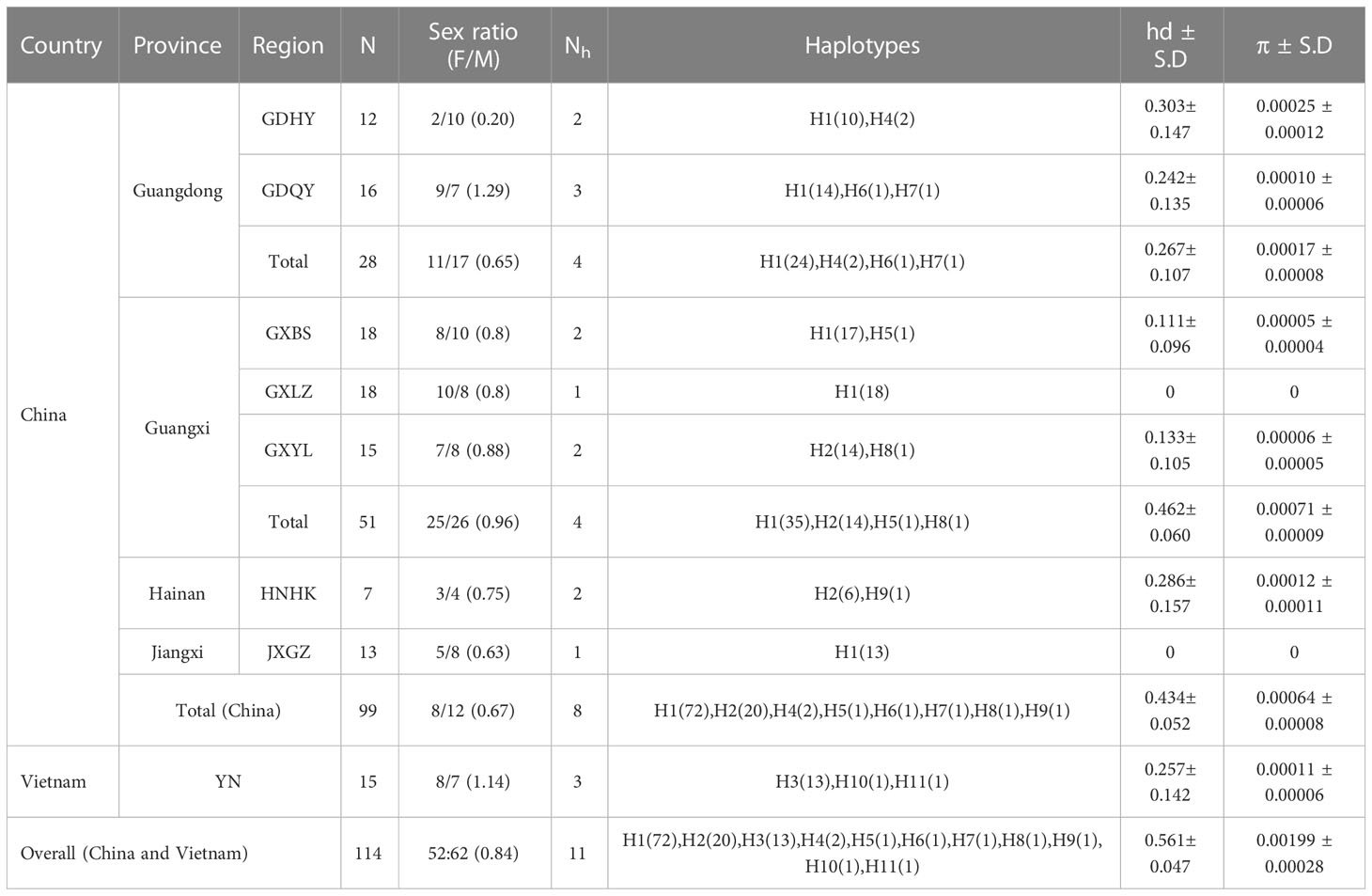
Table 1 Summary of samples and genetic diversity of Mastacembelus armatus (N: number of samples; Nh: number of nrDNA haplotypes; hd: haplotype diversity; π: nucleotide diversity; F/M: ratio of the number of females to males).
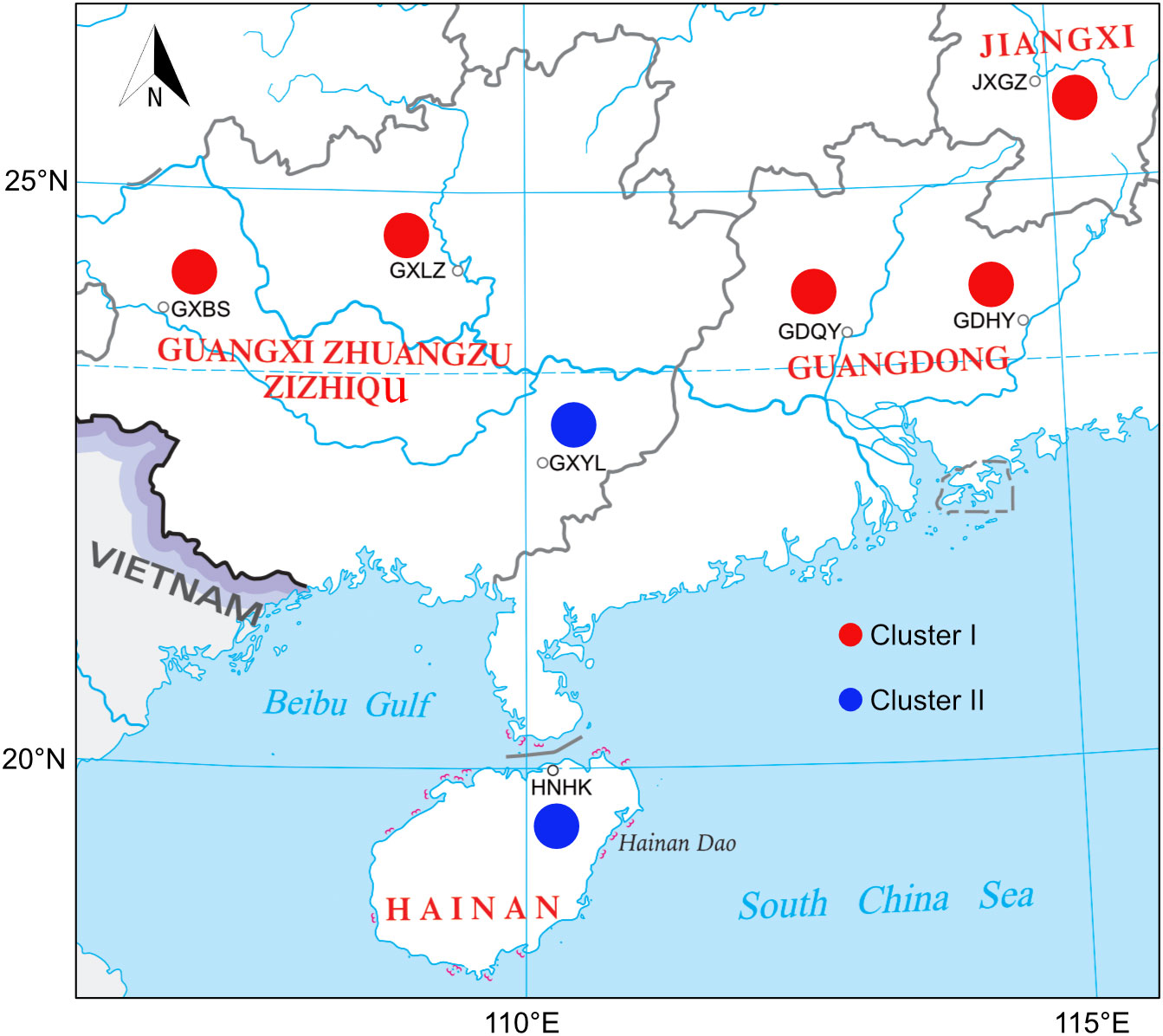
Figure 1 Distribution of sampling sites of Mastacembelus armatus populations in China. Region abbreviations follow those in Table 1. Pie charts represent the Cluster frequency of nrDNA haplotypes belong to in each region; the colour of each Cluster is consistent with Figures 2, 3.
DNA extraction and sequencing
A 30-40 mg fin clip was collected and preserved in 95% ethanol at -20°C for later genomic sequencing. Both DNA sequencing and assembly were performed by Science Corporation of Gene (SCGene) Co., Ltd, Guangzhou city, Guangdong Province, China. Total genomic DNA was extracted with a Tissue DNA Kit (OMEGA E.Z.N.A) following the manufacturer’s protocol. The quality and quantity of genomic DNAs were determined by 0.8% agarose gel electrophoresis and NanoDrop 2000 spectrometer (Thermo Scientific, Waltham, MA, USA). High-quality genomic DNAs were used to construct a paired-end sequencing library with an insert size of 450 bp. The library was then sequenced on an Illumina novaseq 6000 (Illumina, USA) high-throughput sequencing platform in accordance with the manufacturer’s instructions.
Sequence assembly
Adaptors and low-quality reads were filtered using Trimmomatic v0.39 (Bolger et al., 2014), resulting the raw reads, which number were between 3,876,630 and 51,352,359. Paired-end reads of 2 × 150 bp were generated, and the quality threshold was set to Q20. Qualified reads were then compared by BWA (Li and Durbin, 2009) employing setting of 0 match and 0 gap. Afterwards, the obtained reads were assembled using SOAPdenovo (Luo et al., 2012). To verify the correctness of the assembly, assembled whole nrDNA sequences were amplified and sequenced by Sanger sequencing. The annotation of assembled nrDNAs was performed using blastn in NCBI with closely related and well-annotated sequences, manually verified afterwards. Finally, respective region sequences were generated, including 18S, ITS1, 5.8S, ITS2 and 28S.
Data analysis
Standard diversity indices, including number of haplotypes (Nh), haplotype diversity (hd) and nucleotide diversity (π), were calculated using DnaSP v 5.10 (Librado and Rozas, 2009). A TCS network was constructed with PopArt (Clement et al., 2002; Leigh and Bryant, 2015) to investigate genealogical relationships among populations inferred from the concatenated sequences of 18S and ITS2. Hierarchical analysis of molecular variance (AMOVA) was performed to detect genetic variations within and among different regions using Arlequin v 3.5 (AMOVA; Excoffier and Lischer, 2010), with statistical significance determined by 1,000 permutations. To quantify the genetic dissimilarity between populations, population pairwise genetic distances (FST) were also calculated by Arlequin v 3.5 (Excoffier and Lischer, 2010). The genetic distance among different populations was used to construct the UPGMA tree in MEGA X (Kumar et al., 2018).
Results
Characteristics of nrDNA
Variations of the length of either whole nrDNA sequences or respective region sequences were slight, see details in Table 2. To be specific, the length of 18S and 5.8S of all individuals were all the same, with 1,840 bp and 154 bp, respectively. Especially, all sequences of 5.8S were completely identical. In addition, there was also very little variation in the length of 28S, with only a 2 bp difference in length. In the 28S alignment of all individuals, 30 variable sites were found, accounting for 0.85%, compared with 3 in 18S (0.16%), 20 in ITS2 (3.37%) and 66 in ITS1 (5.77%). It is worth to note that although the proportion of variable sites in the 28S alignment was low, the majority of variable sites, 21 out of 30, were singletons variable sites. Clearly, ITS1 and ITS2 were regions with greater variability. Furthermore, we found that many indels occurred in the alignment of ITS1, for example, the longest indel was 14 bp in length, located at 1020 nt - 1043 nt. The GC content of whole nrDNA in all populations was similar, between 62.5% and 62.7%, showing a high GC content.
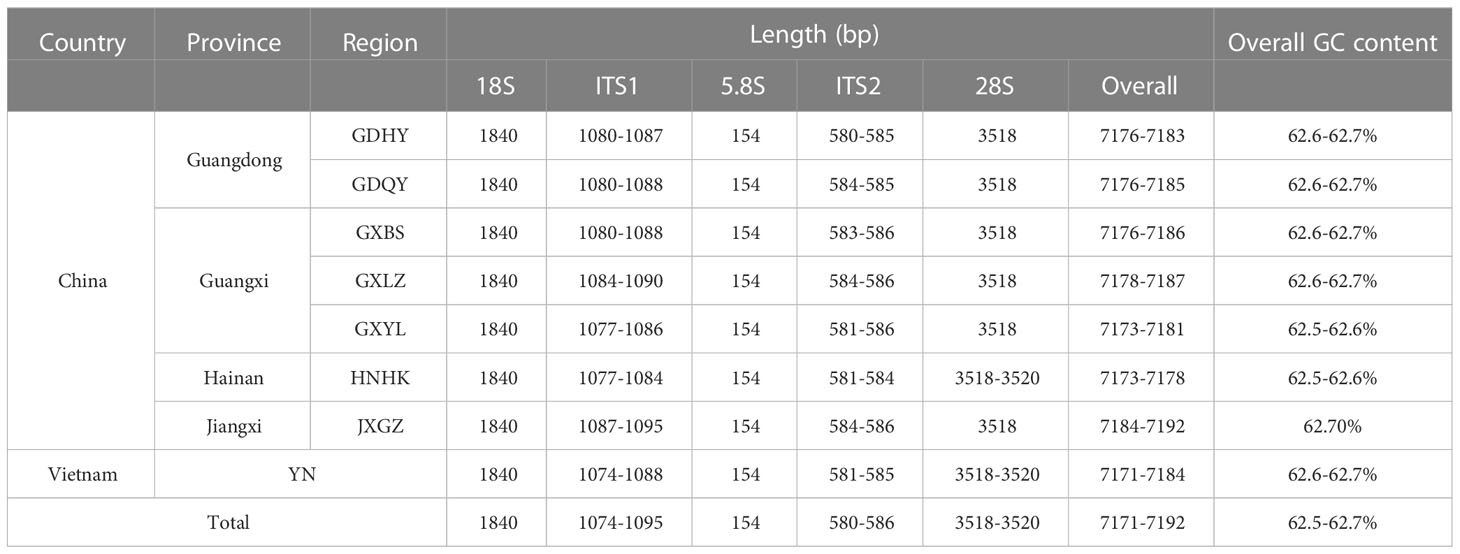
Table 2 The length and GC content of whole nrDNA and respective region of Mastacembelus armatus from eight populations.
More than 5% indels and 5.77% variable sites occurred in the alignment of ITS1, directly affecting the accuracy of subsequent analyses. Additionally, in the alignment of 28S, too many singletons variable sites were detected, which is also thought to negatively impact the molecular analyses, such as phylogenetic analysis and population genetics (Dress et al., 2008; Steenwyk et al., 2020). Besides, the 5.8S sequences of all individuals were the same, we thus decided to use the concatenated sequence of 18S and ITS2 for all later analyses.
Genetic diversity
Eleven nrDNA haplotypes were identified from the concatenated sequences of 18S and ITS2, consistently inferred from DnaSP results and TCS network (Table 1, Figure 2). The overall genetic diversity was relatively high (0.561 ± 0.047) while nucleotide diversity was low (0.00199 ± 0.00028) (Table 1). The highest haplotype diversity was found in GDHY region in China, followed by HNHK, YN, and GDQY region. Most regions harbored more than one haplotype, except GXLZ and JXGZ region. In a word, we found eight haplotypes in China and three haplotypes in Vietnam (Table 1, Figure 2), and H1, H2 and H3 were the main haplotypes, making three different Clusters (Figure 2). Amongst, H1 and H2 were the main haplotypes found in China and H3 in Vietnam. Moreover, in China, the genetic diversity of M. armatus in Guangxi province was the highest (0.462 ± 0.060), consisting of two main haplotypes H1 and H2, while only H1 was detected in Jiangxi Province, predominant H1 in Guangdong Province, and predominant H2 in Hainan Province.
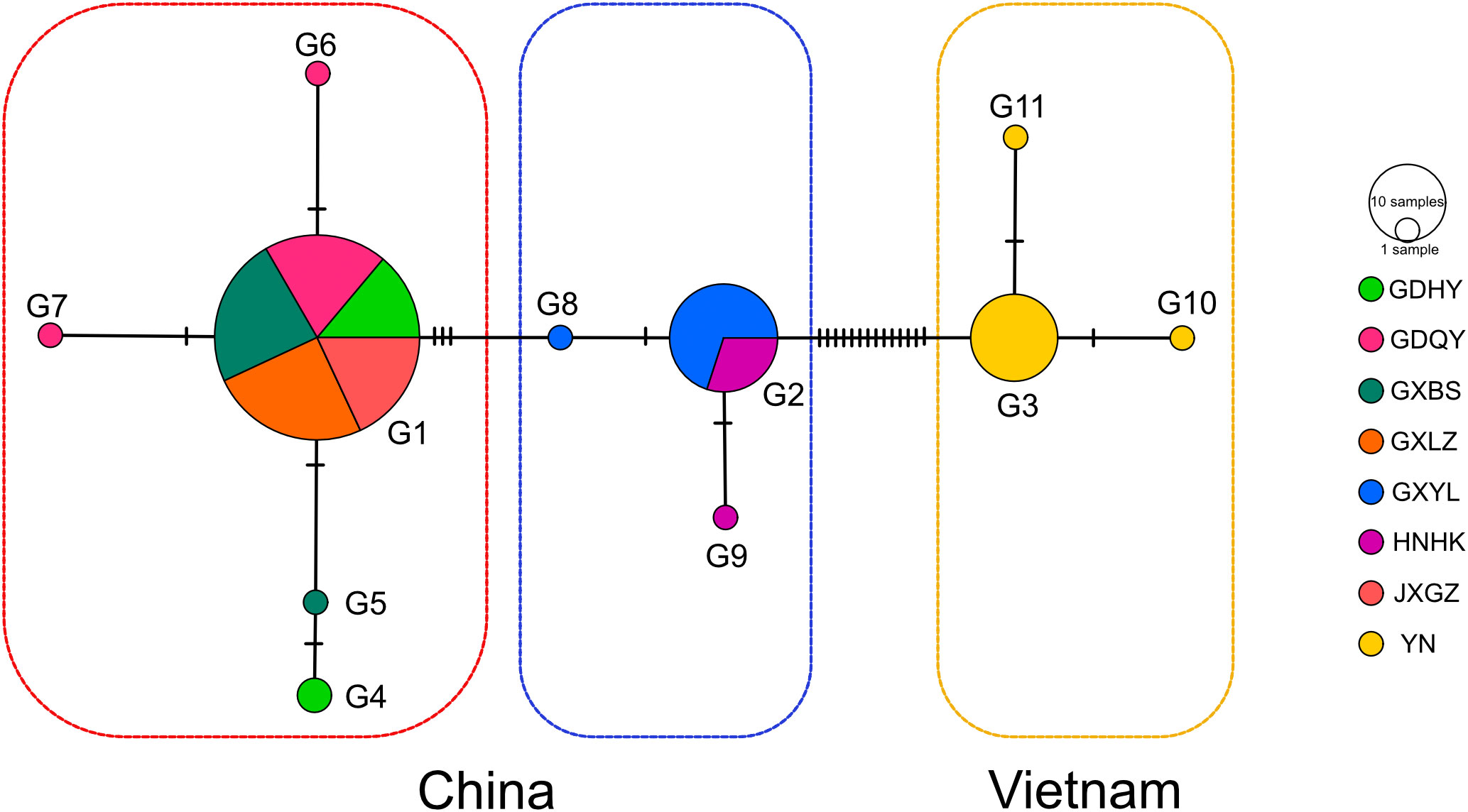
Figure 2 TCS network based on the concatenated sequences of 18S and ITS2 of Mastacembelus armatus. Each tick represents a mutational step. nrDNA haplotypes are named as in Table 1. Circle size is proportional to the haplotype frequency.
Overall, more males were found in our study. In Vietnam, the number of females exceeded males, compared to Chinese populations with dominant males.
Population structure
In general, AMOVA analyses presented that there was high genetic differentiation in overall populations of M. armatus (ΦST = 0.882, p < 0.001), and the largest level of genetic differentiation was found among populations (88.19%) (Table 3). The similar pattern was shown at the province level (ΦST = 0.796, p < 0.001) and at the country level (ΦST = 0.886, p < 0.001) as well (Table 3). Pairwise FST comparisons revealed that most populations were significantly differentiated (Table 4). Particularly, pronounced differences among different provinces were found in a range of FST between 0.210 and 0.970, all being statistically significant (Table 5), in agreement with AMOVA result that most variations were from among provinces (79.59%) at the province level (Table 3). In addition, the UPGMA tree demonstrated that all populations were divided into three groups (Figure 3). Cluster I consisted of all populations from Jiangxi and Guangdong Province and two of three populations of Guangxi Province (GXLZ and GXBS). While the other population of Guangxi Province (GXYL) was clustered with the population from Hainan Province, forming Cluster II, consistent with the low FST value between them, only 0.004. The Vietnam population was a single group, Cluster III. Overall, Cluster I and Cluster II were grouped together, making the Chinese Cluster. Furthermore, the genetic differentiation of Clusters between China and Vietnam was also distinctive, characterized with different nrDNA haplotypes (Table 1, Figure 2) and distinct phylogenetic Clusters (Figure 3), as well as shown in the AMOVA analysis, with 88.59% variances from among countries (Table 3).
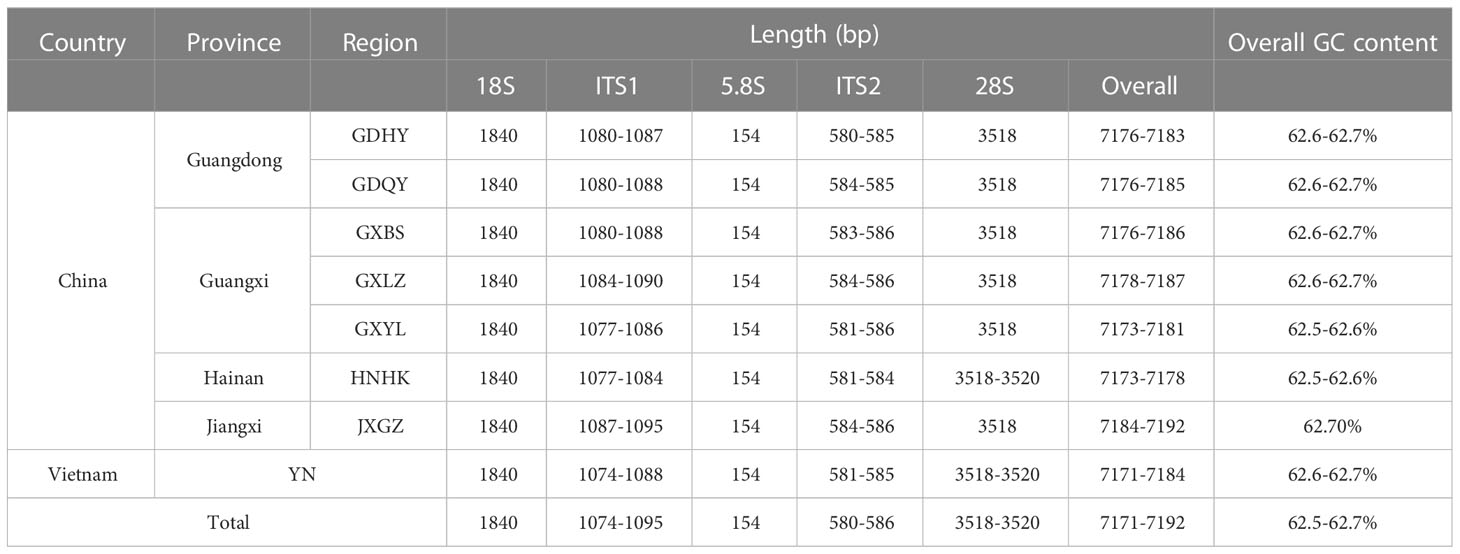
Table 3 Analysis of molecular variance (AMOVA) of Mastacembelus armatus populations at different spatial scale.
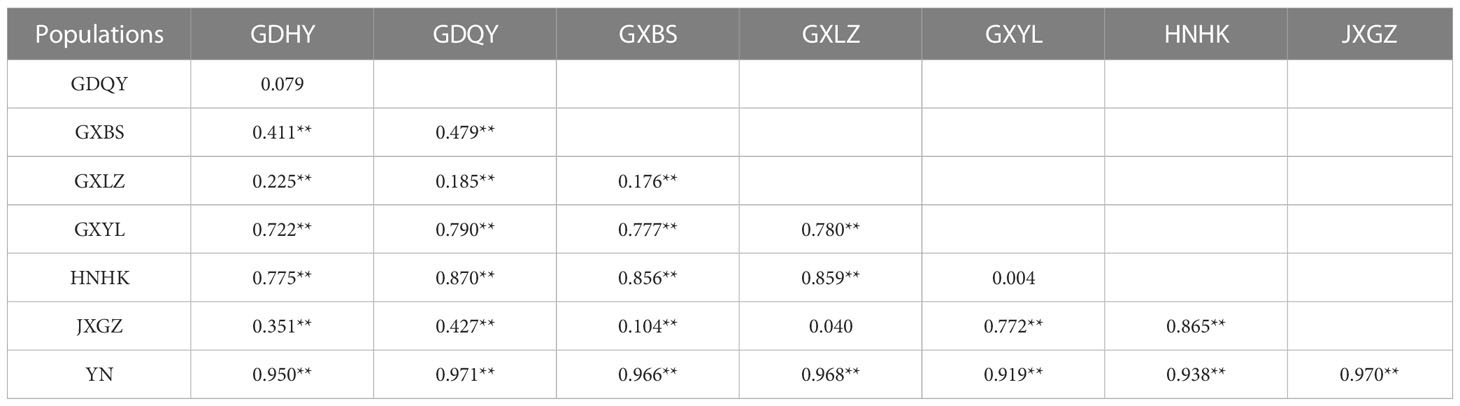
Table 4 Pairwise estimates of genetic differentiation (FST) of Mastacembelus armatus between different populations (**p<0.05).
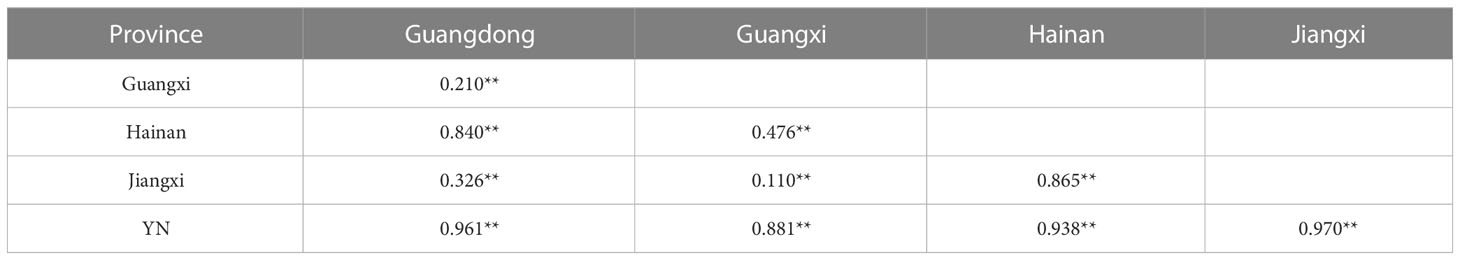
Table 5 Pairwise estimates of genetic differentiation (FST) of Mastacembelus armatus between different provinces (**p<0.05).
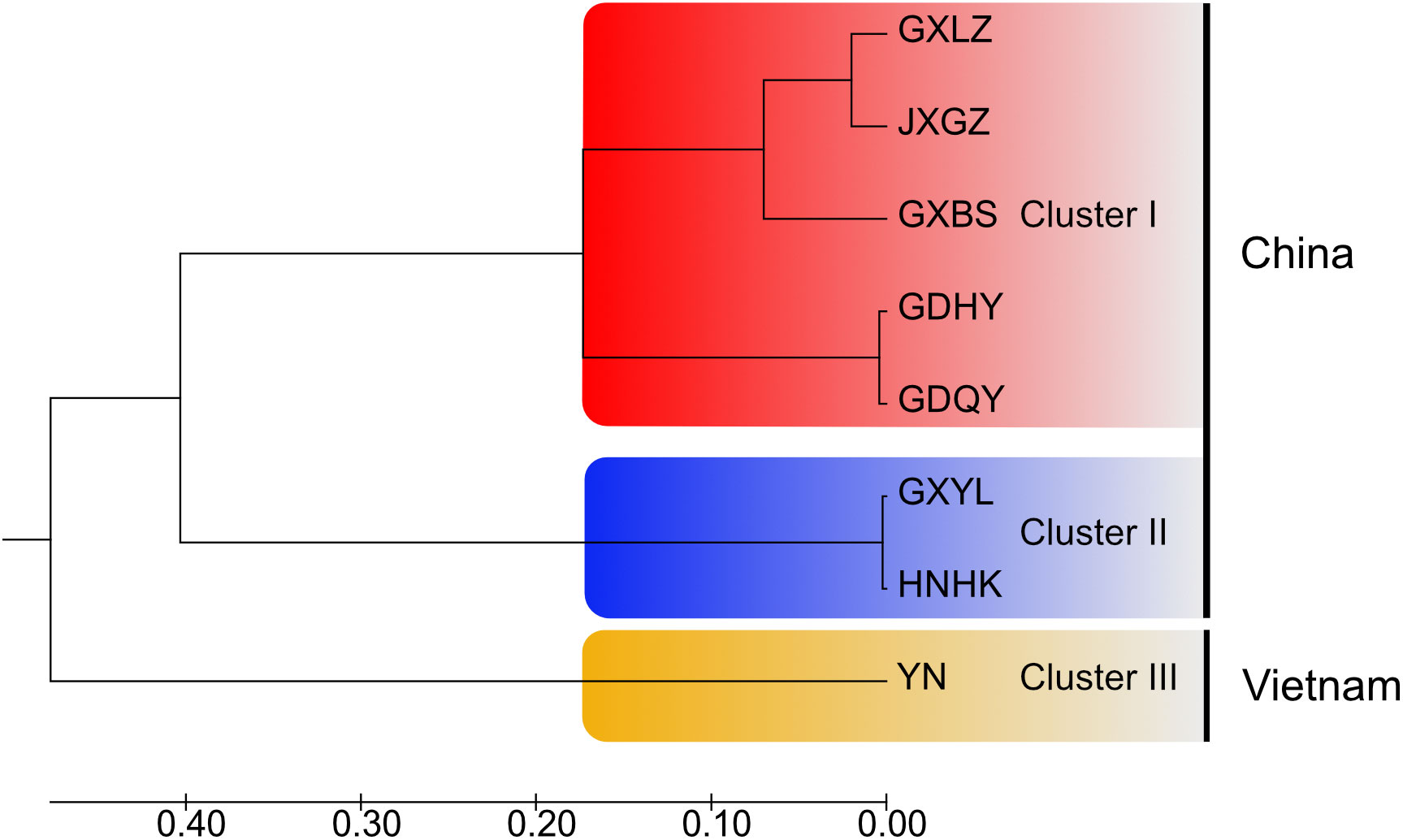
Figure 3 UPGMA dendrogram of eight populations of Mastacembelus armatus based on pairwise genetic distances between populations.
Discussion
Genetic diversity and population structure
Previous population studies (Wang et al., 2012; Zou, 2013; Chen, 2014; Yang et al., 2016; Lin, 2017; Jiang, 2018; Gao et al., 2022) all showed high genetic diversity of M. armatus populations in China, but the haplotype diversity in our result (hd=0.434) is lower than that in other population studies, for example, Wang et al. (2012), Jiang (Jiang, 2018) and Gao et al. (2022) reported the haplotype diversity in China was 0.965, 0.768 and 0.895, respectively. This may be due to different markers employed and the small sample size in our study compared to other studies. Additionally, the overall genetic diversity of M. armatus populations in this study was over 0.5, showing a relative high diversity (Grant and Bowen, 1998). In general, combing our data and literature data, the genetic diversity of M. armatus maintains at a relatively high level, although the size of natural population is declining as reported (Hossain et al., 2012; Rahman et al., 2016; Xue, 2018). Furthermore, genetic differentiation between most populations is pronounced, regardless of spatial scale. Guangxi Province harbored the highest genetic diversity. Amongst, GXLZ population and GXBS population were grouped together, in agreement with low FST value between them. GXYL population, nevertheless, was in a distinct cluster, either shown in TCS network or UPGMA tree. This suggests not only high genetic diversity but also high genetic differentiation among Guangxi Province populations. Surprisingly, GXYL population was clustered together with HNHK population. Further, the low FST value between Hainan Province and the mainland means that gene flow occurred between them, despite the fact that Hainan Province is an isolated island and separated from the mainland by sea waters. In contrast, most Chinese population studies revealed that the population from Hainan Province was genetically partitioned with the mainland populations (Yang et al., 2016; Lin, 2017; Jiang, 2018). Gene flow between the population of Hainan Province and the mainland, revealed by nuclear makers in our study, is congruent with the latest research of Gao et al. (2022) discovered by mitochondrial markers. This gene flow is probably as a consequence of geological changes during glacial periods, when the exposed continental shelf connected Hainan and the mainland China (Sun et al., 2000; Voris, 2000). However, Guangxi Province and Vietnam possess completely different nrDNA haplotypes, despite their proximity. This is mainly because that there are no rivers connecting Guangxi Province and Vietnam.
Sex ratio
It is well known that the population size is closely related with sex ratio, and an unbalanced sex ratio may significantly reduce the effective size of populations (Dubreuil et al., 2010). We checked the sex of each individual after sampling, and we found that all sampling sites in China are male dominated, except GDQY. However, Vietnam is female dominated. In fact, the sex ratio of M. armatus in the nature is on debate. One research reported female dominance trend (Panikkar et al., 2013), while the other one showed an equal proportion of male and female (Serajuddin and Pathak, 2012). We need more natural population data to clarify the sex ratio of M. armatus in the future, in order to provide more references for the further biological conservation. In addition, we discovered that the sex ratio of M. armatus is very unbalanced during the artificial breeding process, characterized with significant female dominance. For example, the proportion of females can reach 86.33% in the report of Xue et al. (2021b). This suggests that there are differences and difficulties in sexual differentiation of M. armatus, which may also occur in nature. At present, although some studies have been done on the sex differentiation of M. armatus, most focus on the development of sex markers (Xue et al., 2020; Xue et al., 2021b) and Y chromosome differential (Xue et al., 2021a). The underlying sex determination mechanisms are still unclear, which also make challenges to investigate the sex ratio of M. armatus.
Biological conservation
The biological conservation of species heavily depends on their genetic diversity. Understanding intraspecific genetic diversity and differentiation can help us take scientific and effective measures to protect threatened or endangered animals. Combing the results of our study with other previous studies, we found that the present genetic diversity of M. armatus populations is not low, indicating that it is not too late to take action to protect them, so that their genetic diversity can remain high. For example, precisely constructing more reserves for M. armatus based on the distribution of different genotypes/haplotypes, strengthening the investigation and protection of M. armatus in the isolated island, Hainan Province, which may provide crucial clues for their expansion, and exploring artificial breeding techniques to better conserve the germplasm resource.
Conclusion
In conclusion, we researched the genetic diversity and population structure of M. armatus in China and Vietnam, based on nuclear ribosomal DNA markers. The genetic diversity and differentiation of M. armatus populations were at a relatively high level according to the data from this study and previous studies. Three Clusters were classified according to the genetic distance between each population, characterized with two Clusters in China and a distinct Cluster in Vietnam. In particular, we found that gene flow occurred between an isolated island and the mainland China based on the low FST value between them.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, OP847241 - OP847354.
Ethics statement
The study was approved by the Laboratory Animal Ethics Committee of Pearl River Fisheries Research Institute, CAFS (number: LAEC-PRFRI-20201219).
Author contributions
Conceptualization: XM. Resources: YY, YWa, YWu, YL, CL, ZJ. Methodology: XM, YY. Investigation: YY, YWa, YWu, YL, CL, ZJ. Analysis: YY, YWa, XM. Writing – original draft: YY and YWa wrote the manuscript. Writing – review and editing: all authors. Funding acquisition: XM and YWu. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Rural Revitalization Strategy Special Provincial Organization and Implementation Project Funds (2022RS-2-1), Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Team (2022KJ150), China-ASEAN Maritime Cooperation Fund (CAMC-2018F), and National Freshwater Genetic Resource Center (FGRC18537).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ağdamar S., Tarkan A. S. (2019). High genetic diversity in an invasive freshwater fish species, Carassius gibelio, suggests establishment success at the frontier between native and invasive ranges. Zoologischer Anzeiger 283, 192–200. doi: 10.1016/j.jcz.2019.10.002
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Chen T. T. (2014). Population differentiation and phylogeography of Mastacembelus armatus in southern China based on COI and d-loop (Master dissertation, south China normal university, China).
Clement M., Snell Q., Walke P., Posada D., Crandall K. (2002). TCS: estimating gene genealogies. In Proceedings of the 16th International Parallel and Distributed Processing Symposium, International (IEEE Computer Society) 3, 184.
Dhole J., Tambe D., Chavan R. (2011). Histopathological study of Mastacembelus armatus (Leceped, 1800) infected with tapeworm from osmanabad district (MS) India. Recent Research in Science and Technology 3, 3.
Dress A. W., Flamm C., Fritzsch G., Grünewald S., Kruspe M., Prohaska S. J., et al. (2008). Noisy: identification of problematic columns in multiple sequence alignments. Algorithms Mol. Biol. 3 (1), 1–10. doi: 10.1186/1748-7188-3-7
Dubreuil M., Riba M., González-Martínez S. C., Vendramin G. G., Sebastiani F., Mayol M. (2010). Genetic effects of chronic habitat fragmentation revisited: strong genetic structure in a temperate tree, Taxus baccata (Taxaceae), with great dispersal capability. Am. J. Bot. 97 (2), 303–310. doi: 10.3732/ajb.0900148
Duong T. Y., Tran L. V. D., Nguyen N. T. T., Jamaluddin J. A. F., Azizah M. N. S. (2020). Unravelling taxonomic ambiguity of the Mastacembelidae in the Mekong delta (Vietnam) through DNA barcoding and morphological approaches. Tropical Zoology 33 (2).
Excoffier L., Lischer H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol. Ecol. Resour. 10 (3), 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fan H. P., Qiu M. L., Zhong Q. F., Xue L. Z., Qin Z. Q. (2018). Comparison of nutritional component of wild and cultured Mastacembelus armatus in different growth stages. J. Anhui Agric. Sci. 46 (8), 92–96.
Gao S., Li Y. F., li J., Chen W. T. (2022). Genetic structure and demographic history of Mastacembelus armatus in southern China. South China Fisheries Sci. 19, 1–8. doi: 10.12131/20220200
Garcia G., Pereyra S., Gutierrez V., Oviedo S., Miller P., Domingo A. (2015). Population structure of Squatina guggenheim (Squatiniformes, squatinidae) from the south-western Atlantic ocean. J. Fish Biol. 86 (1), 186–202. doi: 10.1111/jfb.12560
Grant W. A. S., Bowen B. W. (1998). Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J. Heredity 89 (5), 415–426. doi: 10.1093/jhered/89.5.415
Gupta S., Banerjee S. (2016). Food, feeding habit and reproductive biology of tire-track spiny eel (Mastacembelus armatus): A review. J. Aquaculture Res. Dev. 7 (5), 429. doi: 10.4172/2155-9546.1000429
Han C., Li Q., Lin J., Zhang Z., Huang J. (2018). Characterization of complete mitochondrial genomes of Mastacembelus erythrotaenia and Mastacembelus armatus (Synbranchiformes: Mastacembelidae) and phylogenetic studies of Mastacembelidae. Conserv. Genet. Resour. 10 (3), 295–299. doi: 10.1007/s12686-017-0807-0
Han C., Li Q., Liu J., Hao Z., Huang J., Zhang Y. (2019). Characterization, evolution, and expression analysis of TLR7 gene subfamily members in Mastacembelus armatus (Synbranchiformes: Mastacembelidae). Dev. Comp. Immunol. 95, 77–88. doi: 10.1016/j.dci.2019.02.002
Han C., Li Q., Zhang Z., Huang J. (2017). Characterization, expression, and evolutionary analysis of new TLR3 and TLR5M genes cloned from the spiny eel Mastacembelus armatus. Dev. Comp. Immunol. 77, 174–187. doi: 10.1016/j.dci.2017.08.007
Hossain M. Y., Hossen M. A., Yahya K., Ahmed Z. F., Sarder M. R. I., Islam M. A., et al. (2015). Threatened fishes of the world: Mastacembelus armatus (Lacepede 1800) (Synbranchiformes: Mastacembelidae). Croatian J. Fisheries 73 (3), 137–139. doi: 10.14798/73.3.820
Hossain M. Y., Rahman M. M., Jewel M. A. S., Ahmed Z. F., Ahamed F., Fulanda B., et al. (2012). Conditions-and form-factor of the five threatened fishes from the jamuna (Brahmaputra river distributary) river, northern Bangladesh. Sains Malaysiana 41 (6), 671–678.
IUCN (2019). IUCN red list of threatened species. Version 2019, (IUCN Red List of Threatened Species)3.
IUCN Bangladesh. (2000). Red book of threatened fishes of Bangladesh (Bangladesh: IUCN Bangladesh, Dhaka) 116.
Javed M., Usmani N. (2016). Accumulation of heavy metals and human health risk assessment via the consumption of freshwater fish Mastacembelus armatus inhabiting, thermal power plant effluent loaded canal. SpringerPlus 5 (1), 1–8. doi: 10.1186/s40064-016-2471-3
Jiang X. L. (2018). Genetic diversity and phylogeography of different populations of Mastacembelus armatus in southern China and its adjacent areas (Master dissertation, guangzhou university, China).
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547. doi: 10.1093/molbev/msy096
Leigh J. W., Bryant D. (2015). POPART: Full-feature software for haplotype network construction. Methods Ecol. Evolution. 6 (9), 1110–1116. doi: 10.1111/2041-210X.12410
Li H., Durbin R. (2009). Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 25 (14), 1754–1760. doi: 10.1093/bioinformatics/btp324
Li Q., Xu R., Shu H., Chen Q., Huang J. (2016). The complete mitochondrial genome of the zig-zag eel Mastacembelus armatus (Teleostei, Mastacembelidae). Mitochondrial DNA Part A 27 (1), 330–331. doi: 10.3109/19401736.2014.892102
Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 (11), 1451–1452. doi: 10.1093/bioinformatics/btp187
Lin T. T. (2017). Isolation of microsatellite markers and population genetic diversity analysis in Mastacembelus armatus (Master dissertation, guangzhou university, China).
Lin W. Q., Liao X. P., Chen T., Xu Z. S., Liang W. L., Su Y. L. (2016). Research on artificial propagation techniques for Mastacembelus armatus. Ocean and Fishery 7, 50–53.
Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1 (1), 2047–217X. doi: 10.1186/2047-217X-1-18
Mladineo I., Šegvić-Bubić T., Stanić R., Desdevises Y. (2013). Morphological plasticity and phylogeny in a monogenean parasite transferring between wild and reared fish populations. PloS One 8 (4), e62011. doi: 10.1371/journal.pone.0062011
Oliveira C., Torres R. A., Favorito S., Foresti F. (1997). Cytogenetic studies of Mastacembelus armatus (Pisces, Mastacembelidae). Cytobios 83–89.
Pandey M., Pandey A. K., Mishra A., Tripathi B. D. (2017). Assessment of metal bioaccumulation in Mastacembelus armatus (eel) and exposure evaluation in human. Environ. Nanotechnology Monit. Manage. 7, 103–109. doi: 10.1016/j.enmm.2017.02.002
Panikkar P., Khan M. F., Sharma A. P., Jha B. C., Vijaykumar M. E. (2013). Index of relative importance of diet components in Mastacembelus armatus (Lacepède 1800) from karapuzha reservoir, wayanad, kerala, India. Indian J. Fisheries 60 (1), 37–40.
Presa P., Pardo B. G., Martínez P., Bernatchez L. (2002). Phylogeographic congruence between mtDNA and rDNA ITS markers in brown trout. Mol. Biol. Evol. 19, 2161–2175. doi: 10.1093/oxfordjournals.molbev.a004041
Rahman M. M., Ali M. R., Sarder M. R. I., Mollah M. F. A., Khan N. S. (2016). Development of sperm cryopreservation protocol of endangered spiny eel, Mastacembelus armatus (Lacepede 1800) for ex-situ conservation. Cryobiology 73 (3), 316–323. doi: 10.1016/j.cryobiol.2016.10.004
Serajuddin M., Pathak B. C. (2012). Study of reproductive traits of spiny eel, Mastacembelus armatus (Mastacembeliforms) from kalinadi-a tributary of the gangesriver basin, India. Res. J. Biol. 2 (05), 145–150.
Shu H., Jiang X. L., Yang H. Q., Lin T. T., Zhou H. Q., Zhang M. Q., et al. (2017). Analysis of morphological variations among seven wild populations of Mastacembelus armatus in south China area. J. Guangzhou Univ. (Natural Sci. Edition) 16 (3), 8–14.
Steenwyk J. L., Buida T. J. III, Li Y., Shen X. X., Rokas A. (2020). ClipKIT: A multiple sequence alignment trimming software for accurate phylogenomic inference. PloS Biol. 18 (12), e3001007. doi: 10.1371/journal.pbio.3001007
Sun X., Li X., Luo Y., Chen X. (2000). The vegetation and climate at the last glaciation on the emerged continental shelf of the south China Sea. Palaeogeography Palaeoclimatology Palaeoecol. 160 (3-4), 301–316. doi: 10.1016/S0031-0182(00)00078-X
Thapliyal M., Sati B. K., Thapliyal A. (2020). Assessing impact of dams on genetic diversity of native fish Mastacembelus armatus in river yamuna using mitochondrial DNA cytochrome-b sequences as a molecular marker. Environ. Conserv. J. 21 (1&2), 39–47. doi: 10.36953/ECJ.2020.211205
Voris H. K. (2000). Maps of pleistocene sea levels in southeast Asia: shorelines, river systems and time durations. J. Biogeography 27 (5), 1153–1167. doi: 10.1046/j.1365-2699.2000.00489.x
Wang F., Huang X. L., La D., Zhao J. (2012). Research on population genetics and phylogeography of mastacembelus armatus populations in south China area. In Proceedings of the 2012 Symposium of the Ichthyology Branch of the Chinese Society of Zoology, Lanzhou, Chinese Society for Oceanology and Limnology, Chinese Ichthyological Society, 23.
Wu Y., Liang Z. Q., Li C. W., Liu L., Chen X. Y., Liu Q. M. (2010). Analysis and evaluation of nutritional components in the muscle of two kinds of Mastacembelus. Acta Nutrimenta Sin. 32 (5), 499–502. doi: 10.13325/j.cnki.acta.nutr.sin.2010.05.012
Xue L. (2018). Effects of exogenous hormone, temperature and parental weight on artificial induced spawning and incubation of Mastacembelue armatus. Acta Hydrobiologica Sin. 42 (2), 333–341. doi: 10.7541/2018.042
Xue L., Gao Y., Wu M., Tian T., Fan H., Huang Y., et al. (2021a). Telomere-to-telomere assembly of a fish y chromosome reveals the origin of a young sex chromosome pair. Genome Biol. 22 (1), 1–20. doi: 10.1186/s13059-021-02430-y
Xue L. Z., Guo X. F., Zhou Y. L., Wang Z. W., Fan H. P., Li D. P., et al. (2020). Screening and characterization of sex-specific markers by 2b-RAD sequencing in zig-zag eel (Mastacembelus armatus) with implication of XY sex determination system. Aquaculture 528, 735550. doi: 10.1016/j.aquaculture.2020.735550
Xue L., Jia D., Xu L., Huang Z., Fan H., Chen B., et al. (2021b). Bulk and single-cell RNA-seq reveal the sexually dimorphic expression pattern of dmrtb1 in zig-zag eel (Mastacembelus armatus). Aquaculture 545, 737194. doi: 10.1016/j.aquaculture.2021.737194
Yang H. Q., Li Q., Shu H., Yue L., Lin T. T., Liu. Y. B. (2016). Genetic diversity of Mastacembelus armatus in southern China and surrounding areas based on ISSR analysis. Acta Hydrobiologica Sin. 40 (1), 63–70. doi: 10.7541/2016.9
Zhou H. Q., Li F., Shu H., Zhong D. M., He P. Y., Huang X. Q., et al. (2019). Analysis on morphological indexes and discrimination of Male and female Mastacembelus armatus. J. Guangdong Ocean Univ. 39 (1), 1–6. doi: 10.3969/j.issn.1673-9159.2019.01.001
Keywords: genetic diversity, populations, mastacembelus armatus, southern China, Vietnam
Citation: Yang Y, Wang Y, Wu Y, Liu Y, Liu C, Jiang Z and Mu X (2023) Population genetics of zig-zag eel (Mastacembelus armatus) uncover gene flow between an isolated island and the mainland China. Front. Mar. Sci. 10:1100949. doi: 10.3389/fmars.2023.1100949
Received: 17 November 2022; Accepted: 21 February 2023;
Published: 06 March 2023.
Edited by:
Wei Liu, Qilu University of Technology (Shandong Academy of Sciences), ChinaReviewed by:
Victor Hugo Valiati, University of the Rio dos Sinos Valley, BrazilKhaled Mohammed-Geba, University of Maryland, College Park, United States
Copyright © 2023 Yang, Wang, Wu, Liu, Liu, Jiang and Mu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xidong Mu, muxd@prfri.ac.cn
 Yexin Yang1,2
Yexin Yang1,2  Xidong Mu
Xidong Mu