Mechanisms of carbon dioxide detection in the earthworm Dendrobaena veneta
- 1Department of Biology, Wake Forest University, Winston-Salem, NC, United States
- 2Department of Biology, Boston University, Boston, MA, United States
- 3Department of Biological Sciences, Kean University, Union, NJ, United States
Introduction: Carbon dioxide (CO2) is a critical biological signal that is noxious to many animals at high concentrations. The earthworm Dendrobaena veneta lives in subterranean burrows containing high levels of CO2 and respires through its skin. Despite the ecological and agricultural importance of earthworms, relatively little is known about how they make decisions in their environment, including their response to elevated levels of CO2.
Methods: To examine CO2 detection in this species, we designed the exudate assay, in which we placed an earthworm in a sealed container, exposed it to varying concentrations of CO2 for one minute, and recorded the amount of exudate secreted. Because earthworms excrete exudate in response to noxious stimuli, we hypothesized that the amount of exudate produced was proportional to the amount of irritation. We repeated these experiments after treatment with several blockers for molecules with potential involvement in CO2 detection, including carbonic anhydrases, guanylate cyclase, TRPA1, ASICs, and OTOP channels. We also confirmed the presence of homologous transcripts for each of these gene families in an epithelial transcriptome for D. veneta. Additionally, since organisms often detect CO2 levels indirectly by monitoring the conversion to carbonic acid (a weak acid), we used the exudate assay to evaluate aversion to additional weak acids (formic acid, acetic acid, and propionic acid).
Results: Earthworms excreted significantly more exudate in response to CO2 in a dosage-dependent manner, and this response was muted by the general carbonic anhydrase inhibitor acetazolamide, the carbonic anhydrase IX/XII inhibitor indisulam, the calcium channel blocker ruthenium red, the sodium channel blocker amiloride, and the acid-sensing ion channel blocker diminazene aceturate.
Discussion: These data provide evidence of the role of carbonic anhydrase and epithelial sodium channels in earthworm CO2 detection, establish that, similar to other subterranean-dwelling animals, earthworms are extremely tolerant of CO2, and contribute to our understanding of the mechanisms used by earthworms to detect and react to weak acids in their environment.
Introduction
As a major byproduct of cellular respiration, CO2 is pervasive in most ecosystems and may indicate the presence of other living organisms. It is thus a critical signaling molecule that is often attractive or aversive depending on the organism and concentration. Fruit flies, for example, avoid CO2 released by neighboring stressed flies (Suh et al., 2004), while mosquitoes are attracted to CO2 emitted by their hosts (Spanoudis et al., 2020). At higher concentrations, CO2 can be noxious, and may result in hypercapnia, hypoxia, or anesthesia (Cummins et al., 2020). Given its biological prevalence, potential health risks, and environmental relevance, it is critical to understand the mechanisms by which organisms detect CO2.
Important decomposers in some ecosystems and invasive species in others, earthworms play critical roles in environmental health and agriculture. In many ecosystems, earthworms are a key component of soil fertility, influencing soil turnover, soil aeration, and nutrient availability (Edwards, 2004). Despite their essential environmental role, we know little about what chemicals attract and repel earthworms nor the mechanisms by which they detect those chemicals (Silver et al., 2019; Reed et al., 2021). One such chemical is carbon dioxide (CO2). The current concentration of CO2 in air is approximately 0.04% (Amundson and Davidson, 1990; Scott, 2011). Earthworms live in burrows up to three meters deep and may encounter CO2 concentrations of 0.04% to 13.0% (Amundson and Davidson, 1990). Some subterranean organisms, such as naked mole rats, have unique adaptations that allow them to tolerate normally noxious concentrations of CO2 (Shams et al., 2005; Fang et al., 2014); earthworms may have similar adaptations.
The ubiquitous presence and broad importance of CO2 has produced multiple detection pathways across organisms (Figure 1A). Most mechanisms require carbonic anhydrase, which catalyzes the reversible conversion of carbon dioxide into a bicarbonate ion and a proton, reacting with water from extracellular fluid (Lindskog, 1997; Figure 1A). Carbonic anhydrase is found across animals, plants, and microorganisms, includes three independently evolved isozyme families, and is essential in many biological functions including metabolism, cellular transport, and acid-base balance (Henry, 1996; Banerjee and Deshpande, 2016). The multiple isoforms of α-carbonic anhydrases, the family found in animals, are labeled CAI to CAXV and are differentially expressed among tissues and cell types (Tarun et al., 2003). Mechanisms of CO2 detection may respond to CO2 directly, to bicarbonate ions, or to protons from the carbonic anhydrase reaction (Makino et al., 2019; Figure 1A).
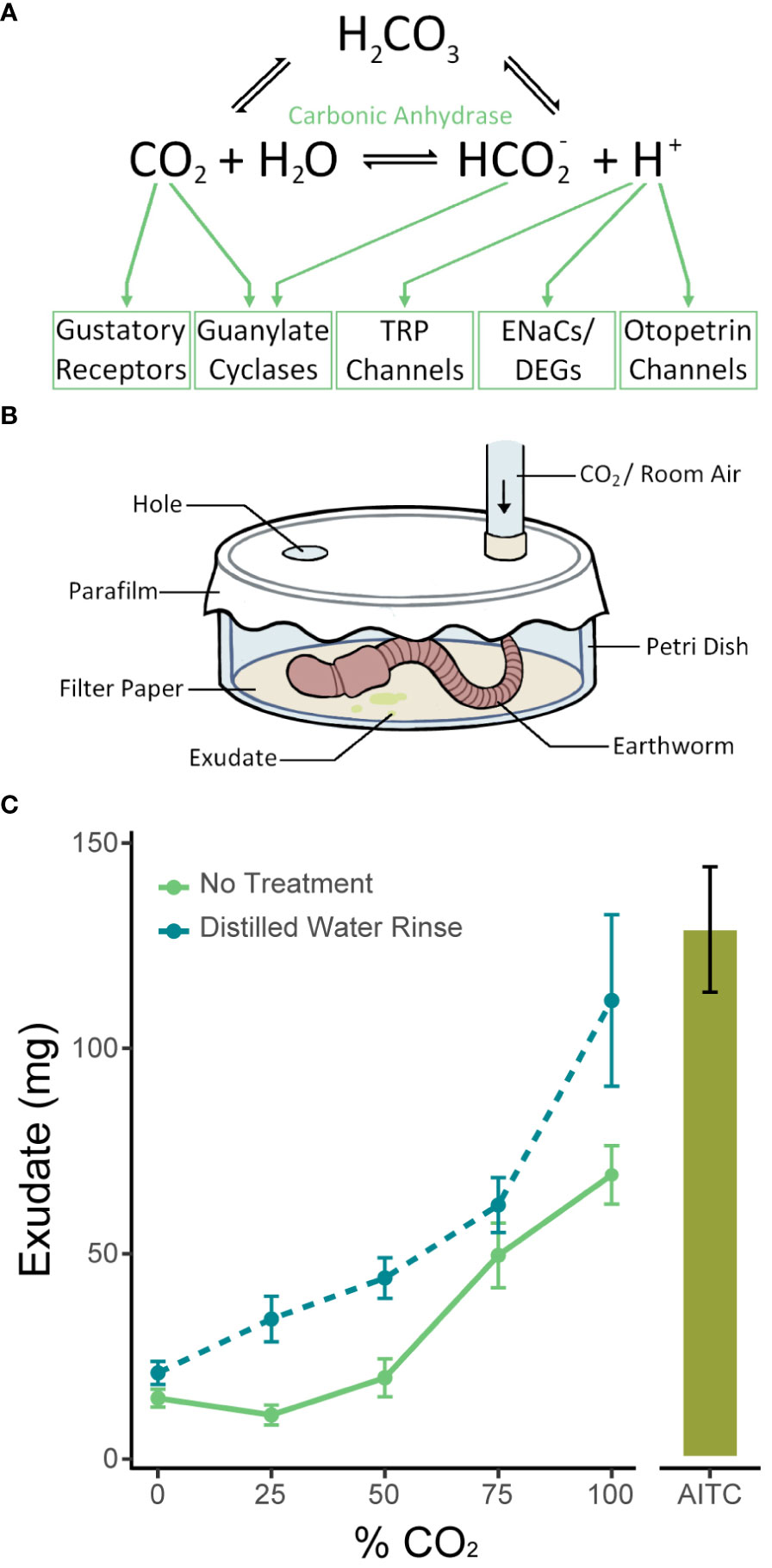
Figure 1 Mechanism of CO2 detection and the exudate assay. (A) The chemical reaction CO2 + H2O ←→ HCO2- + H+ is catalyzed by carbonic anhydrase. The green arrows point to the potential mechanisms by which CO2 and its products could be detected. (B) Diagram of the exudate assay. Earthworms were placed in a sealed container filled with a volatile or gaseous chemical for one minute, and the amount of exudate produced in that time was recorded. (C) Earthworms excreted significantly more exudate in response to increasing concentrations of CO2 (p<0.0001, n=12-20 one-way ANOVA). Earthworms also excreted significantly more exudate in response to CO2 if they were rinsed in distilled water prior to the assay (p<0.0001, two-way ANOVA). The response to air flowed over filter paper containing 500 µl of 500 mM AITC, a known noxious stimulus, is shown for the purposes of assay validation. Graphed values are means ± SEM.
D. veneta lives in burrows several cm below the ground and detects chemicals in the soil using sensory cells on its epithelium, either grouped into epithelial sensory organs (ESOs) or found alone as solitary chemoreceptor cells (Hess, 1925; Csoknya et al., 2005; Kiszler et al., 2012). The cells project to the ventral nerve cord and are categorized into five types based on fine structure (Csoknya et al., 2005; Kiszler et al., 2012). These organs are more abundant on the anterior segments and are the likely candidates for the cellular receptors responsible for CO2 and weak acid detection. However, this study is the first attempt to determine the molecular identity of the genes responsible for CO2 detection in these tissues.
Here we investigate CO2 and weak acid detection in the European Nightcrawler Dendrobaena veneta (previously known as Eisenia hortensis) using known mechanisms from other species as a template. Ionotropic gustatory receptors (GRs), acid-sensing ion channels (ASICs), guanylate cyclases (GCs), otopetrin channels, and transient receptor potential ankyrin 1 (TRPA1) channels have all been implicated in the detection of CO2 or weak acids like carbonic acid in other animals. There is some evidence of these mechanisms in D. veneta. Earthworms likely have functioning TRPA1 channels, as they show behavioral aversion to TRPA1 activators (Silver et al., 2018). We expected high concentrations of CO2 to be noxious to earthworms as in other species, and we hypothesized that carbonic anhydrases, guanylate cyclases, TRPA1, ENaCs, and OTOPs may be involved with CO2 detection in earthworms based on the analysis of a D. veneta epithelial transcriptome presented in this study.
Materials and methods
Earthworms
D. veneta earthworms were housed in plastic bins (approximate volume 40L) filled half-way with moist topsoil covered with paper. The bins were maintained at room temperature with two light sources placed above the bins on a 12-12 light-dark cycle. The worms were watered with 250 ml of water poured over the newspaper twice a week and fed with approximately 5 g of “Purina Worm Chow” once a week. Only adult earthworms, identified by their visible clitellum, were used in experiments.
CO2 aversion
Earthworms excrete exudate in response to noxious stimuli (Heredia et al., 2008). Given that high concentrations of CO2 are aversive in most species, we predicted that the degree of exudate production would reflect CO2 detection and relative aversiveness. To perform the exudate assay, we first gently rinsed the earthworms in tap water and placed them in glass containers with moist paper towels to deprive them of soil and allow their digestive tract to empty. After 24 hours, a worm was placed in a glass petri dish with filter paper at the bottom and folded up the sides, and the dish was sealed with parafilm. The parafilm was punctured twice, and the dish was then saturated with air containing some percentage of CO2 through a nozzle inserted through the parafilm for one minute. The filter paper was weighed before and immediately after each experiment to calculate the weight of exudate excreted (Figure 1B).
Using this method, we tested responses to atmospheric CO2, 25% CO2, 50% CO2, 75% CO2, and 100% CO2 and analyzed the data with a one-way ANOVA and a TukeysHSD. Varying percentages of CO2 were obtained by combining different flow rates of room air and 100% CO2 using rotameters and a bubble flow meter for a final flow rate of approximately 32 ml/s for all trials. In this assay, 0% CO2 represents room air, which typically has 400-1,000 ppm CO2 (approximately 0.04%), 25% CO2 includes approximately 250,525 ppm CO2, 50% includes approximately 500,350 ppm CO2, 75% includes approximately 750,175 ppm CO2, and 100% includes approximately 1,000,000 ppm CO2. These results were compared to treatment with distilled water using a two-way ANOVA and TukeysHSD. All exudate assay data was analyzed and graphed in R (R Core Team, 2021) using the packages tidyverse (Wickham et al., 2019), dplyr (Wickham et al., 2023), and ggplot2 (Wickham, 2016). Figures were then manually edited to improve readability and aesthetics. Relevant code for reproducing these methods is available on Github and data files are available with this publication.
Transcriptomics
The earthworm body is covered in sensory cells, which are more abundant on the rostral segments than on the middle segments (Csoknya et al., 2005; Kiszler et al., 2012). In RNA isolated from epithelium covering both of these areas, RNA from the rostral segment will be enriched for transcripts that code for sensory receptor proteins. Thus, the epithelium from the prostomium (1st segment) and 15th segment posterior to the clitellum (hereafter referred to as the mid-segment) were quickly dissected free from the underlying muscle and flash frozen on glass depression slides placed on dry ice. Once frozen solid, the tissue was transferred into 1.5 mL LoBind microcentrifuge tubes (Eppendorf) where it was crushed with a plastic pestle; both items were maintained on dry ice prior to use to maintain their freezing temperature during this process. Refrigerated TRIzol Solution (1mL, Thermo Fisher Scientific) was added, and tissue was further macerated until no intact fragments were visible. Subsequently, RNA was isolated from the tissue-TRIzol mixture using the manufacturer’s standard chloroform extraction and isopropanol precipitation protocol. RNA concentration and integrity was verified using a bioanalyzer (Agilent). This protocol was developed because the earthworm epithelium was surprisingly durable, and other standard methods for tissue disruption (e.g. tissue homogenizers and column systems) failed to yield enough mRNA for sequencing. We dissected the prostomium and mid-segments from 9 earthworms in total. We combined tissue from 3 individuals before extracting RNA to give us 3 prostomium samples and 3 mid-segment samples.
RNA libraries were prepared from total RNA using the Kapa Stranded mRNA-Seq library prep kit, and 150 bp paired-end sequencing was performed on an Illumina HiSeq 4000 at Duke University’s Center for Genomic and Computational Biology (Durham, NC). Trimmomatic v0.36 (Bolger et al., 2014) was used to filter raw reads and to remove Illumina adaptors (4bp, mean Q30). De novo transcriptome assembly was performed by processing the filtered reads with Trinity v.2.5.1 (Haas et al., 2013), and the resulting transcripts were annotated with Trinotate v.3.2.2 (Bryant et al., 2017) using the recommended settings. To develop gene candidate lists, the resulting Trinotate database was filtered for transcripts that were predicted to code for protein and contained one of the following terms: “carbonic anhydrase,” “gustatory receptor”, “ionotropic”, “acid sensing ion channel,” “ASIC,” “guanylate cyclase,” “otopetrin,” “transient receptor potential,” or “TRPA.” These results were then confirmed and collected into candidate lists via human curation and are provided as used for analysis in Supplemental Material 1. Cladograms depicting the similarity of the predicted protein sequence for each group of candidate transcripts were constructed with R (R Core Team, 2021) using the following packages: ggtree (Yu et al., 2017), msa (Bodenhofer et al., 2015), seqinr (Charif and Lobry, 2007), tidyverse (Wickham et al., 2019). We also conducted a differential expression analysis between RNA extracted from the prostomium and midsegment epithelium using DESeq2 Bioconductor package (Love et al., 2014). Relevant code for reproducing these methods is available on Github and sequencing data has been uploaded to the NCBI sequence read archive.
Pharmacology
We repeated the exudate assay after treatment with the inhibitors and blockers described in Table 1. Inhibitors and blockers were sourced from Tocris Bioscience (Acetazolamide, Diminazene Aceturate, S4, Topiramate) and Sigma-Aldrich (Amiloride, HC030013, Methylene Blue, Ruthenium Red, ZnCl2, U-104). For these treatments, the earthworms were placed in a 10 ml conical tube filled with 5 ml of the blocker prior to the experiment. After 10 minutes, they were removed from the tube and gently dried with a paper towel before being placed in the assay chamber. The TRPA1 activator AITC, which we have previously shown to be aversive in D. veneta (Smith, 2019) and Lumbricus terrestris (Silver et al., 2019), was used as a positive control. In these trials, 500 ul of 500 mM AITC (allyl isothiocyanate) was pipetted onto a strip of filter paper placed in a 10 ml pipette, and air was flown through the pipette into the assay chamber. The data were analyzed with two-way ANOVAs and TukeysHSDs. p-values were adjusted using a Bonferroni correction for the carbonic anhydrase inhibitors and the receptor blockers. We observed no mortality during exudate assay experiments, but we did notice less movement or activity after some treatments; AITC controls allowed us to assess whether treatments with significant results affected exudate production rather than CO2 detection.
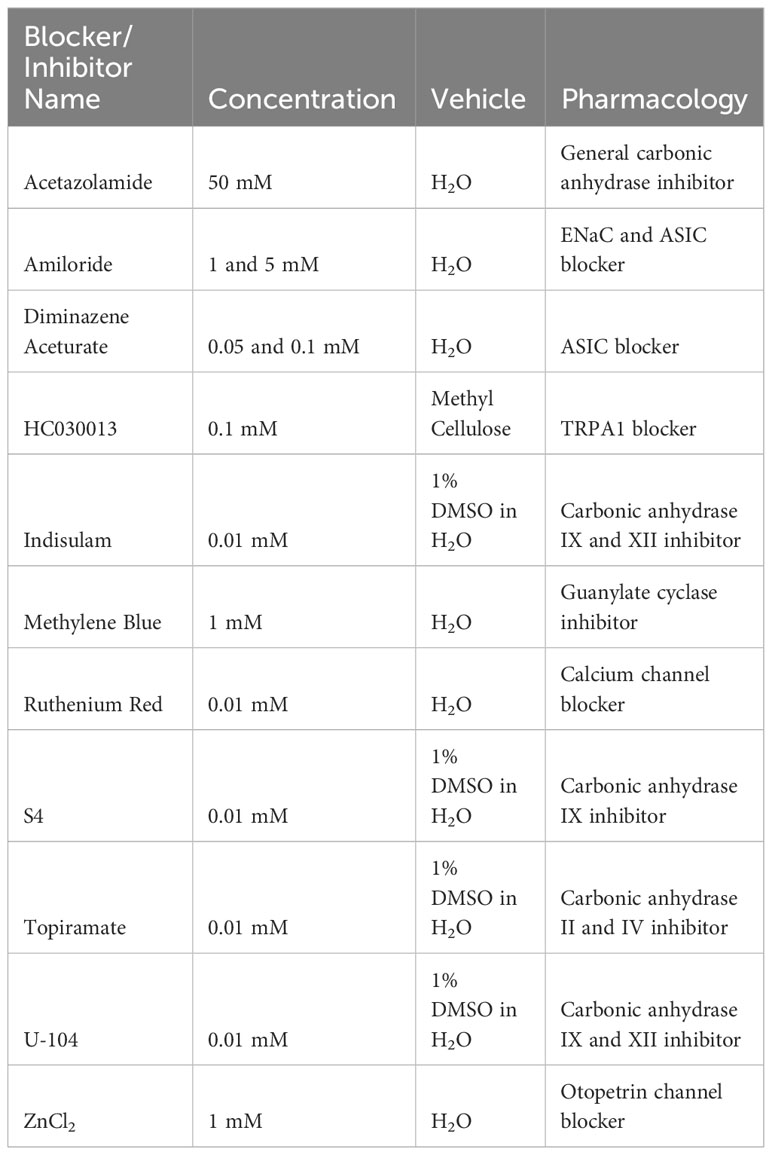
Table 1 Blockers used in the exudate assay with their concentrations, vehicles, and pharmacological interactions of interest.
Organic acids
CO2 detection often relies on the detection of a weak acid (carbonic acid). Therefore, we also studied the responses of earthworms to weak organic acids to assess potential overlapping mechanisms. As detritivores, earthworms likely encounter fermented organic matter that contains organic acids in nature. Formic acid, acetic acid, and propionic acid (Fisher Scientific) were tested at concentrations of 50, 100, 250, 500, 1000 ppb in the exudate assay. For these trials, 250 ul of acid was pipetted onto a strip of filter paper placed in a 10 ml pipette, and air was flown through the pipette into the assay chamber to generate vapor. These trials were repeated at 250 ppb after exposure to the sodium channel and ASIC blocker amiloride using the procedure described above. These data were analyzed using two-way ANOVAs and TukeysHSDs.
Results
CO2 aversion
Earthworms excreted the most exudate in response to 100% CO2. Over the course of the one-minute assay, earthworms first excreted clear liquid, and then a thicker pale green substance. Levels of movement varied between earthworms. Earthworms excreted more exudate with increasing concentrations of CO2 in a dosage-dependent manner (p<0.0001, n=12-20, two-way ANOVA), where concentrations of 0%, 25%, 50%, 75%, and 100% were tested (Figure 1C). Compared to these results, significantly more exudate was excreted when the earthworms were rinsed with distilled water before the experiment (p<0.0001, n=6-12, two-way ANOVA). Washing the worms prior to the experiment likely removes mucus and epithelial surface liquid that has a greater buffering capacity than deionized water, making the earthworm’s epithelium more sensitive to changes in pH.
Transcriptomics
The assembled D. veneta epithelial transcriptome contained 36 protein coding transcripts annotated as carbonic anhydrase (CA) homologs (Figure 2). The presence of CA mRNA supports the idea that these enzymes are involved in an earthworms ability to detect CO2 concentration. Particularly of note are homologs to CAVI, one of the only known secreted CA isoforms which is found in saliva and respiratory epithelium surface liquids of many other species (Leinonen et al., 2004; Fábián et al., 2015). We also queried our transcriptome for genes known to be involved in the detection of CO2 and the various ions associated with carbonic acid and found many.
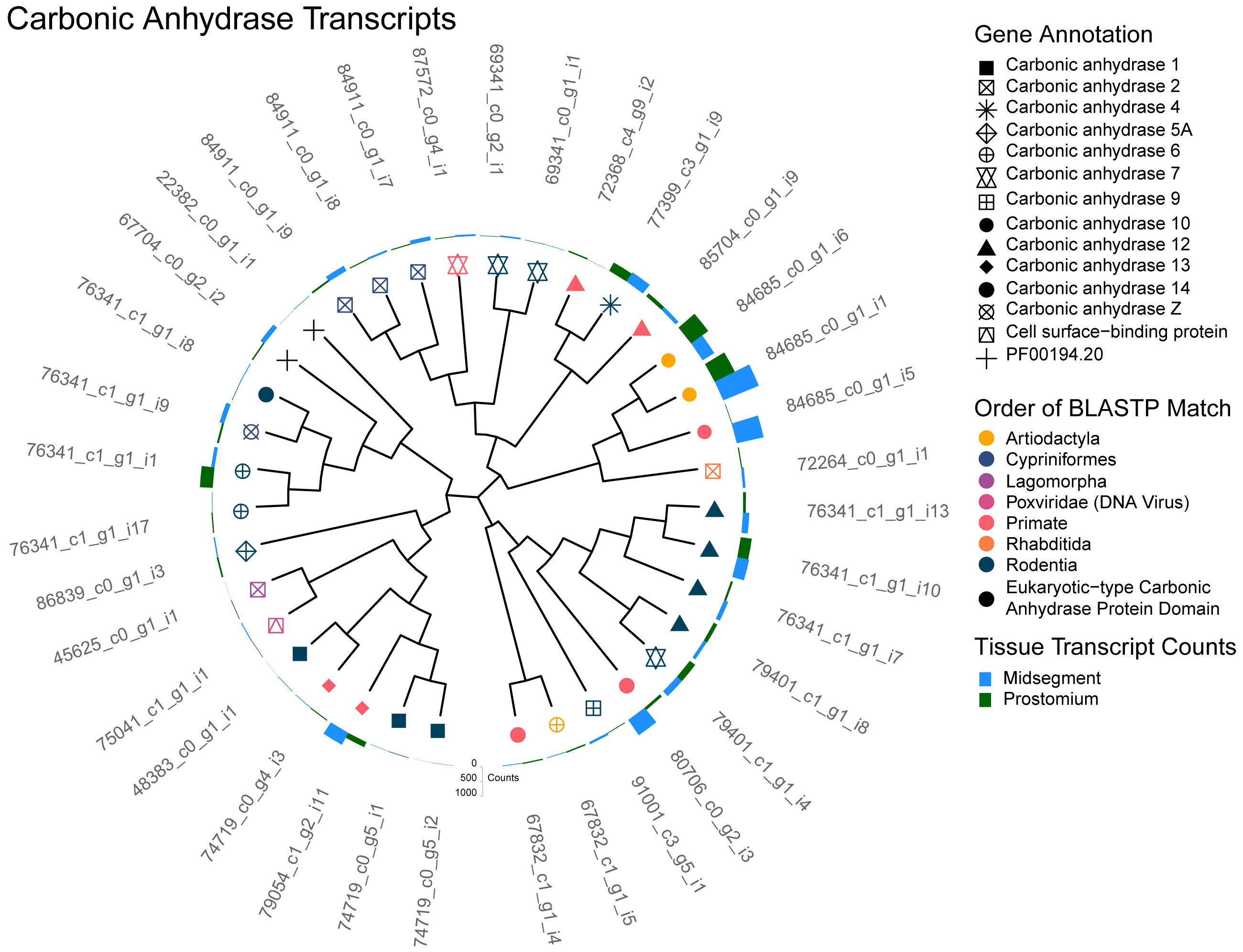
Figure 2 Carbonic anhydrase transcripts. A cladogram depicting the predicted protein sequence similarity between unique transcripts that annotated as carbonic anhydrase (CA). Predicted CA homologs identified by BLASTP are represented as the symbol shape and the order of the homolog as the symbol color. Note that transcript 75041_c1_g1_i1 (pink triangle inside square) showed homology with a viral cell surface-binding protein that contains a protein domain similar to eukaryotic-type carbonic anhydrase. Additionally, two transcripts (black plus sign; 22382_c0_g1_i1 and 67704_c0_g2_i2), did not have a BLASTP homolog but were annotated by Pfam with a eukaryotic-type carbonic anhydrase protein domain (PF00194.20). Total transcript counts from prostomium (green) and midsegment (blue) RNA are depicted as bars at the end of each tree tip.
We did not find any transcripts, however, which annotated as homologs of the ionotropic gustatory receptor family. In insects and many other invertebrates, CO2 directly activates gustatory receptors, a family required for taste and pheromone detection (Kwon et al., 2007; Ning et al., 2016; Chu et al., 2020). C. elegans also detects CO2 directly with isolated chemosensory BAG neurons (Smith et al., 2013). To ensure that our filtering criteria were not artifactually eliminating this gene family, we filtered our annotated transcriptome for any occurrence of the term “gustatory” but did not prefilter for protein-coding transcripts. We still found no transcripts annotated as belonging to this gene family. Automated gene annotation pipelines can undercount ionotropic sensory receptors (Agnihotri et al., 2016; McKenzie et al., 2016; McKenzie and Kronauer, 2018), but it would be uncommon for there to be no evidence of transcripts corresponding to ionotropic gustatory receptors if they were enriched in the epithelial tissues we sequenced.
We also filtered our transcriptome for any BLASTP annotations that contained the term “ionotropic” to check for the presence of ionotropic receptors (IRs) in the ionotropic glutamate receptor family. If they were missing entirely, it could indicate a technical problem with either the capture of ionotropic transcripts during the sequencing protocol or the annotation of those transcripts after sequencing, and might raise the prospect of a similar issue with the GR receptor family. If any were present, it would have been of interest because IR64a has been implicated as a CO2 and weak acid sensor in flies (Ai et al., 2010; Ai et al., 2013). We found 18 protein-coding ionotropic receptor transcripts, 17 of which were annotated as homologs to vertebrate glutamate receptors (15 kainate, 1 delta, and 1 NMDA); and one of which was annotated as a homolog to Drosophila IR25a (S1, transcript ID: 91317_c0_g6_i8). Although IR25a is a co-receptor implicated in attracting flies to low levels of CO2 (approximately 3%), this pathway has been demonstrated as separate from the pathways that facilitate avoidance of high concentrations of CO2 in flies (van Breugel et al., 2018). Additionally, knockdown of IR25a in flies did not have an effect on those animals’ response to weak acids (Ai et al., 2013). Since our bioinformatics methods were validated by detecting these ionotropic receptors but no gustatory receptors, we were confident in focusing on other receptor families as candidate CO2 sensors.
In contrast to the absence of gustatory receptors, 29 unique transcripts annotated as guanylate cyclase were identified in the epithelial transcriptome (Figure 3). Bicarbonate ions sometimes activate guanylate cyclases, opening cGMP-sensitive ion channels and increasing cGMP (Sun et al., 2009). This is true in mice, where receptor-type guanylate cyclases GC-G and GC-D are activated by low concentrations of CO2 (Kuhn, 2016), as well as in C. elegans, where chemosensory BAG neurons require the receptor-type guanylate cyclase GCY-9 for their response (Hallem et al., 2011). GCs are more commonly considered to be essential proteins in G-protein-coupled receptor signaling cascades than as direct detectors of CO2 or bicarbonate. However, their presence in the epithelial transcriptome affirms this gene family’s inclusion on our candidate list.
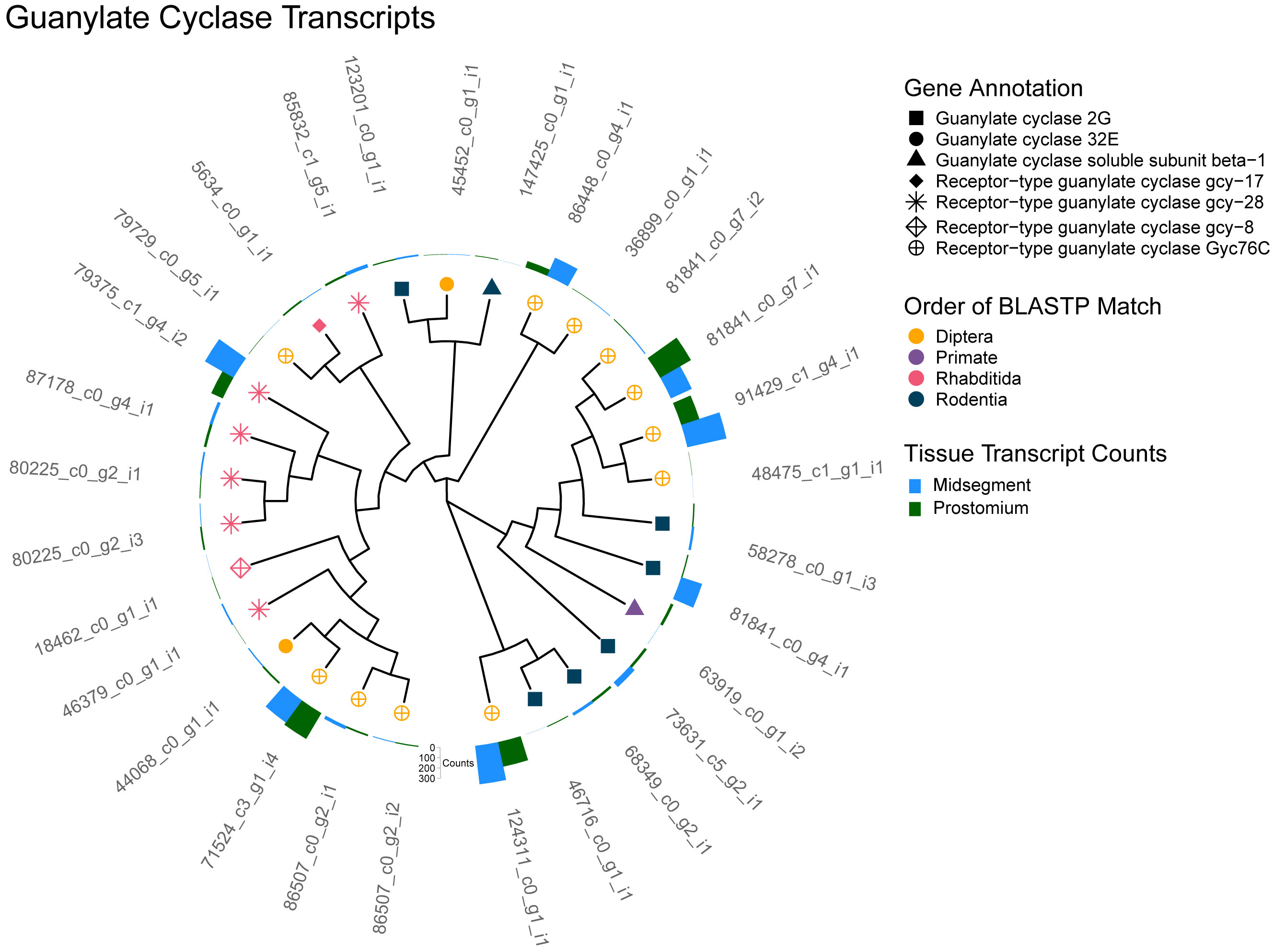
Figure 3 Guanylate Cyclase (GC) transcripts. A cladogram depicting the predicted protein sequence similarity between unique transcripts that annotated as GC. Predicted GC homologs identified by BLASTP are represented as the symbol shape and the order of the homolog as the symbol color. Total transcript counts from prostomium (green) and midsegment (blue) RNA are depicted as bars at the end of each tree tip.
Of the candidate detectors that respond to hydrogen ions produced by the carbonic anhydrase reaction directly, ASICs were the most abundant in the transcriptome with 60 unique transcripts annotated (Figure 4). ASIC channels belong to the Epithelial Na+ channel (ENaC)/degenerin (DEG) channel superfamily. In invertebrates, these channels have been implicated in salt detection, sodium absorption, blood pressure regulation, mechano-transduction, and acid-sensing (Kellenberger and Schild, 2002; Carattino and Montalbetti, 2020). ASIC homologues have been characterized in C. elegans (Rhoades et al., 2019), and there are many DEGs and ENaCs found across invertebrates (Hanukoglu and Hanukoglu, 2016). While ASIC blockage muted CO2 responses in rats (Akiba et al., 2008); the subtype ASIC3 is involved in chemoreception and opens at the physiologically relevant pHs of 7.3 to 6.7 (Li and Xu, 2011; Osmakov et al., 2014), and could thus be involved in CO2 detection. However, loss of function in mice did fully ablate CO2 responses in these animals (Detweiler et al., 2018). One of the 60 ASIC transcripts is annotated as an ASIC3 homolog (Figure 4, S1, 87824_c0_g1_i27). However, the relatively low percent sequence identity (26.5 - 50%, S1) between these transcripts and their ASIC homologs makes us hesitant to narrow this candidate list further based on the Trinotate annotations alone.
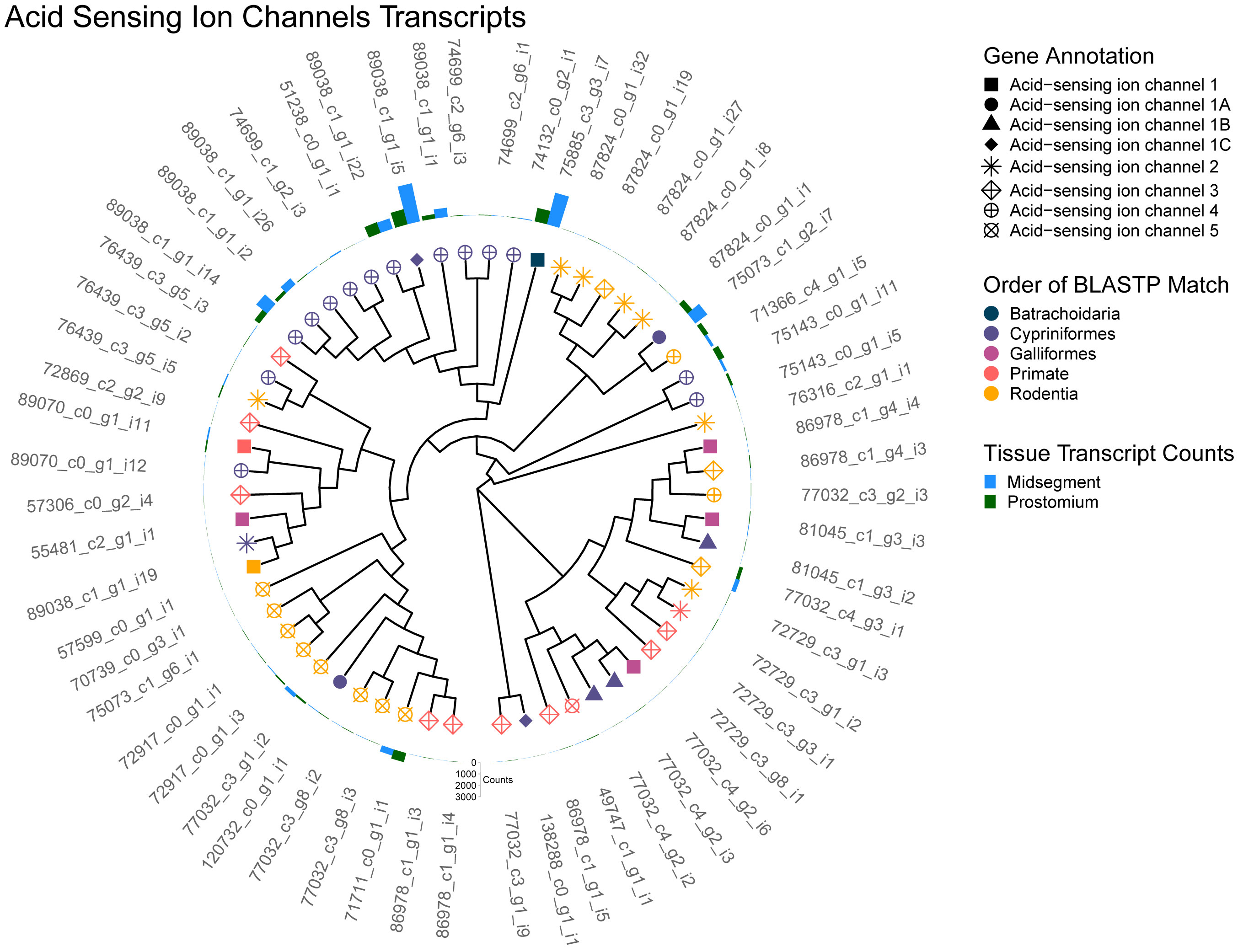
Figure 4 Acid Sensing Ion Channel (ASIC) transcripts. A cladogram depicting the predicted protein sequence similarity between unique transcripts that annotated as ASICs. Predicted ASIC homologs identified by BLASTP are represented as the symbol shape and the order of the homolog as the symbol color. Total transcript counts from prostomium (green) and midsegment (blue) RNA are depicted as bars at the end of each tree tip.
Analysis of the earthworm epithelial transcriptome returned 20 transcripts annotated with an otopetrin protein domain (PF03189.12), and 11 of these were identified as homologs of Drosophila otopetrin (OTOP) via BLASTP (Figures 5, S1). OTOP1 and OTOP3 are conserved across species with related genes in Drosophila (Tu et al., 2018). OTOP1, the proton-sensitive proton channel required for sour taste in mammals (Tu et al., 2018; Teng et al., 2019), may also respond to weak acids such as carbonic acid, a product of the carbonic anhydrase reaction (Teng et al., 2019).
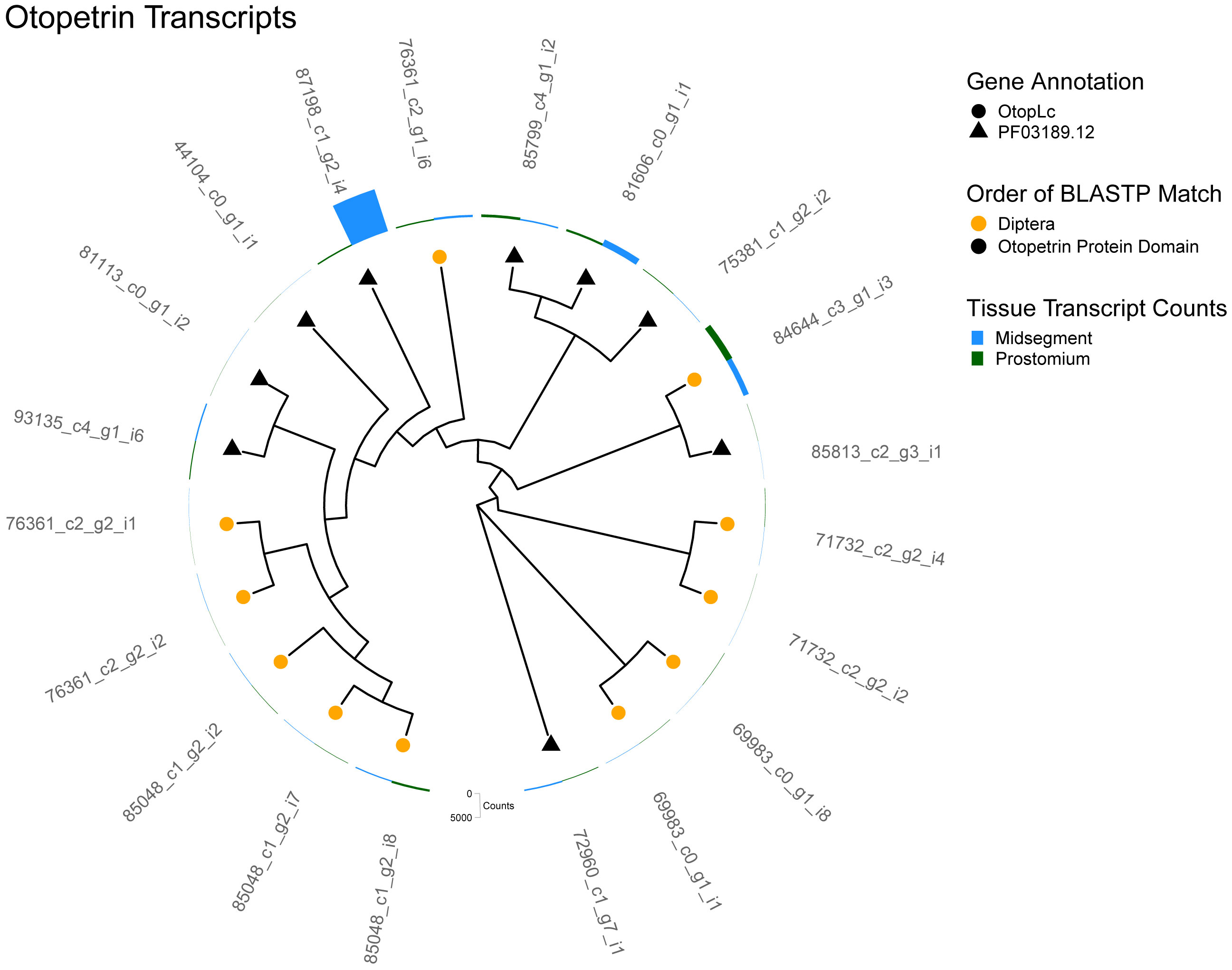
Figure 5 Otopetrin transcripts. A cladogram depicting the predicted protein sequence similarity between unique transcripts that annotated as an otopetrin. Predicted otopetrin homologs identified by BLASTP are represented as the symbol shape and the order of the homolog as the symbol color. Additionally, nine transcripts (black triangle), did not have a BLASTP homolog but were annotated by Pfam with a otopetrin protein domain (PF03189.12). Total transcript counts from prostomium (green) and midsegment (blue) RNA are depicted as bars at the end of each tree tip.
TRPA1 is a calcium channel conserved throughout metazoan life that reacts to irritants, endogenous inflammatory agents, temperature, and weak acids (Bandell et al., 2004; Bautista et al., 2006; Wang et al., 2011). We found 9 transcripts in the earthworm transcriptome that shared homology with TRPA1 (Figure 6). There is evidence in mice and in vitro that the response of sensory neurons to CO2 is mediated by TRPA1 channels activated by intracellular acidification from protons (Takahashi et al., 2008; Wang et al., 2010; Wang et al., 2011). Furthermore, many of the vermifuges used to collect earthworms from the soil are TRPA1 agonists, meaning that there is much behavioral evidence that suggests earthworms have functional TRPA1 channels (Silver et al., 2019).
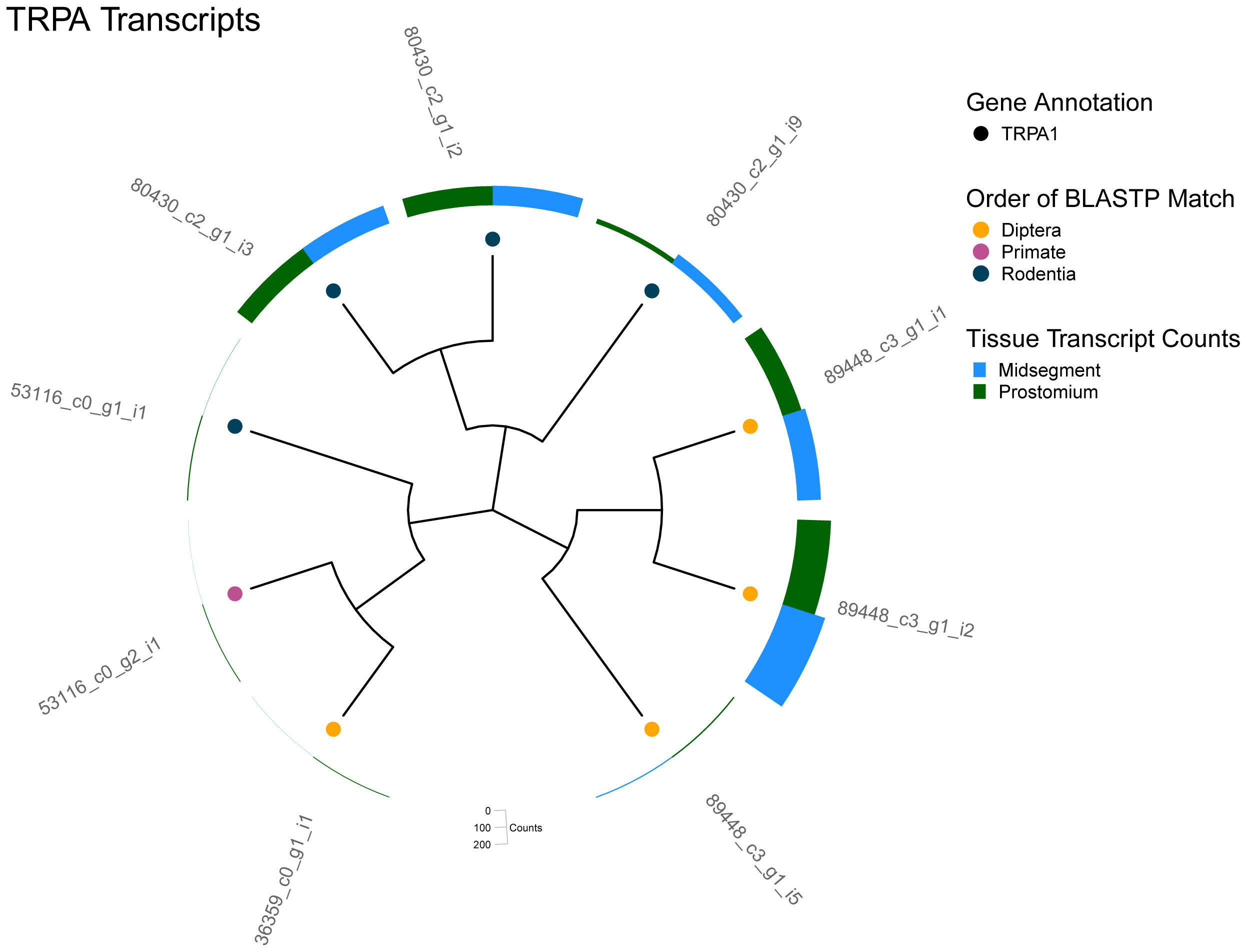
Figure 6 TRPA transcripts. A cladogram depicting the predicted protein sequence similarity between unique transcripts that annotated as TRPA1. Predicted TRPA1 homologs identified by BLASTP are represented as the symbol shape and the order of the homolog as the symbol color. Total transcript counts from prostomium (green) and midsegment (blue) RNA are depicted as bars at the end of each tree tip.
In addition to using a D. veneta epithelial transcriptome to generate a list of candidate genes, we also conducted differential expression analysis between two epithelial tissues. While purported sensory cells are found along the entire length of the earthworm, these cells are more concentrated on the first segments than on segments towards the middle of the body (Csoknya et al., 2005). Since these cells are more abundant near the animal’s head, we hypothesized that transcripts more enriched in this tissue would be more likely to have a sensory function and might provide a clue to the genes responsible for CO2 detection. Transcript counts in both tissues are depicted as bars at the end of each tree branch for CA (Figure 2), GC (Figure 4), ASICs (Figure 3), OTOP (Figure 5), and TRPA1 (Figure 6). While this analysis detected 67 transcripts that were significantly differentially expressed (adjusted p < 0.05) between the two tissues, none of these transcripts were annotated as genes involved in the detection of CO2 or related chemosensory processes (S2). Thus, we chose to test if any of the genes in our candidate list might be involved in the earthworm’s response to noxious levels of CO2.
Pharmacology
The exudate assay was repeated with 0%, 50%, and 100% CO2 after treatment with carbonic anhydrase inhibitors and blockers for receptors potentially involved in CO2 detection (Table 1). As rinsing the earthworms in water increased their sensitivity to CO2 (Figure 1C), all blocker trials were compared to trials with the blocker’s respective vehicle (water, methyl cellulose, or 1% DMSO).
The broad-spectrum CA inhibitor acetazolamide significantly muted the response to CO2 (Figure 7A, p<0.01, n=8-13). This data supports the hypothesis that earthworms are responding to the presence of bicarbonate or hydrogen ions rather than detecting CO2 directly. Attempts to utilize selective CA inhibitors to determine the major CA isoforms involved in the conversion of CO2 were less clear. Indisulam, a blocker of CA IX and CA XII (p<0.05, n=6-8), significantly muted the response to CO2, but U-104, which also blocks CA IV and CA XII, and S4, which also blocks CA IX, resulted in no significant changes (p>0.05, n=6-8). Topiramate, which blocks CA II and CA IV, also did not significantly mute the response to CO2 (p>0.05, n=6-8). The most parsimonious explanation of these results is that one or more isoforms of earthworm CA IX and CA XII have sufficiently diverged from isoforms in other species to make U-104 and S4 ineffective blockers in earthworms.
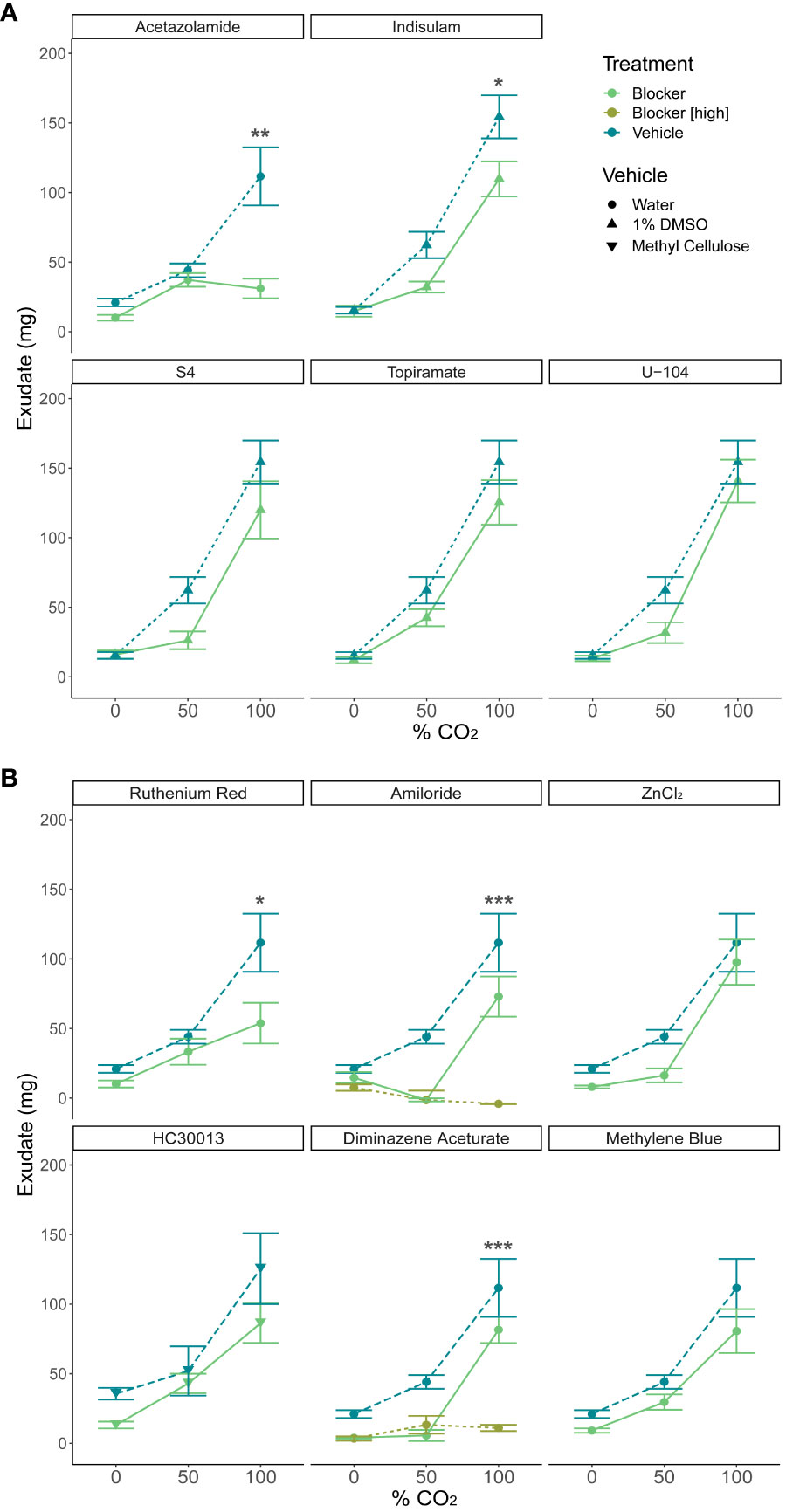
Figure 7 Exudate excretion after treatment with blockers and inhibitors versus treatment with the respective vehicle. (A) The carbonic anhydrase inhibitors acetazolamide (general CA inhibitor, p<0.01, n=8-13) and indisulam (CA IX/XII inhibitor, p<0.05, n=6-8) significantly muted the exudate response by a two-way ANOVA and significant differences were detected via Tukey’s HSD test. S4 (CA IX inhibitor, p>0.05, n=5-8), U-104 (CA IX/XII inhibitor, p>0.05, n=6-8) and topiramate (CA II/IV inhibitor, p>0.05, n=6-8) did not significantly alter the response. (B) Receptor blockers amiloride (ENaC blocker, p<0.0001, n=3-8), diminazene aceturate (ASIC3 blocker, p<0.0001, n=3-8), and ruthenium red (Ca2+ channel blocker, p<0.01, n=5-13) significantly muted the exudate response by a two-way ANOVA, and significant differences were detected via Tukey’s HSD test. HC030013 (TRPA1 blocker, p>0.05, n=6-12), methylene blue (guanylate cyclase inhibitor, p>0.05, n=6-8), and ZnCl2 (OTOP1 blocker, p>0.05, n=6-8) did not significantly alter the response. Graphed values are means ± SEM; * p <0.05, ** p <0.01, *** p <0.001 via Tukey’s HSD test.
Of the receptor blockers, the broad-spectrum ENaC/ASIC blocker amiloride (p<0.0001, n=3-8) and the ASIC3 blocker diminazene aceturate (p<0.0001, n=3-8), and the Ca2+ channel blocker ruthenium red (p<0.05, n=5-13) significantly muted the response to CO2 as detected by two-way ANOVA and Tukeys HSD test (Figure 7B). The TRPA1 channel inhibitor HC030013, the guanylate cyclase inhibitor methylene blue, and ZnCl2 that blocks OTOP1 channels did not significantly alter exudate production (p>.05, n=5-13).
To control for effects of the blockers on the mechanisms of exudate release rather than detection of CO2, the positive control AITC was tested after exposure to acetazolamide, amiloride, diminazene aceturate, indisulam, and HC030013 (Figure 8). Acetazolamide (p>0.05, n=4), 1 mM amiloride (p>.05, n=4), and indisulam (p>0.05, n=5) did not significantly mute the response to AITC by a TukeysHSD test, but HC030013 (p<0.0001, n=7), 5 mM amiloride (p<0.0001, n=4), 0.05 mM diminazene aceturate (p<0.0001, n=5), and 0.1 mM diminazene aceturate (p<001, n=5) did. HC030013 is a TRPA1 blocker and amiloride can act as one at higher concentrations (Eid et al., 2008; Banke, 2011) and predictably blocked AITC detection. The vehicle for AITC, mineral oil, produced no significant response on its own when compared to 0% CO2 (p=1.00, n=4). Taken together these results indicate that Acetazolamide, 1 mM amiloride, and indisulam have no detectable effect on the ability of earthworms to release of exudate when confronted with noxious stimuli.
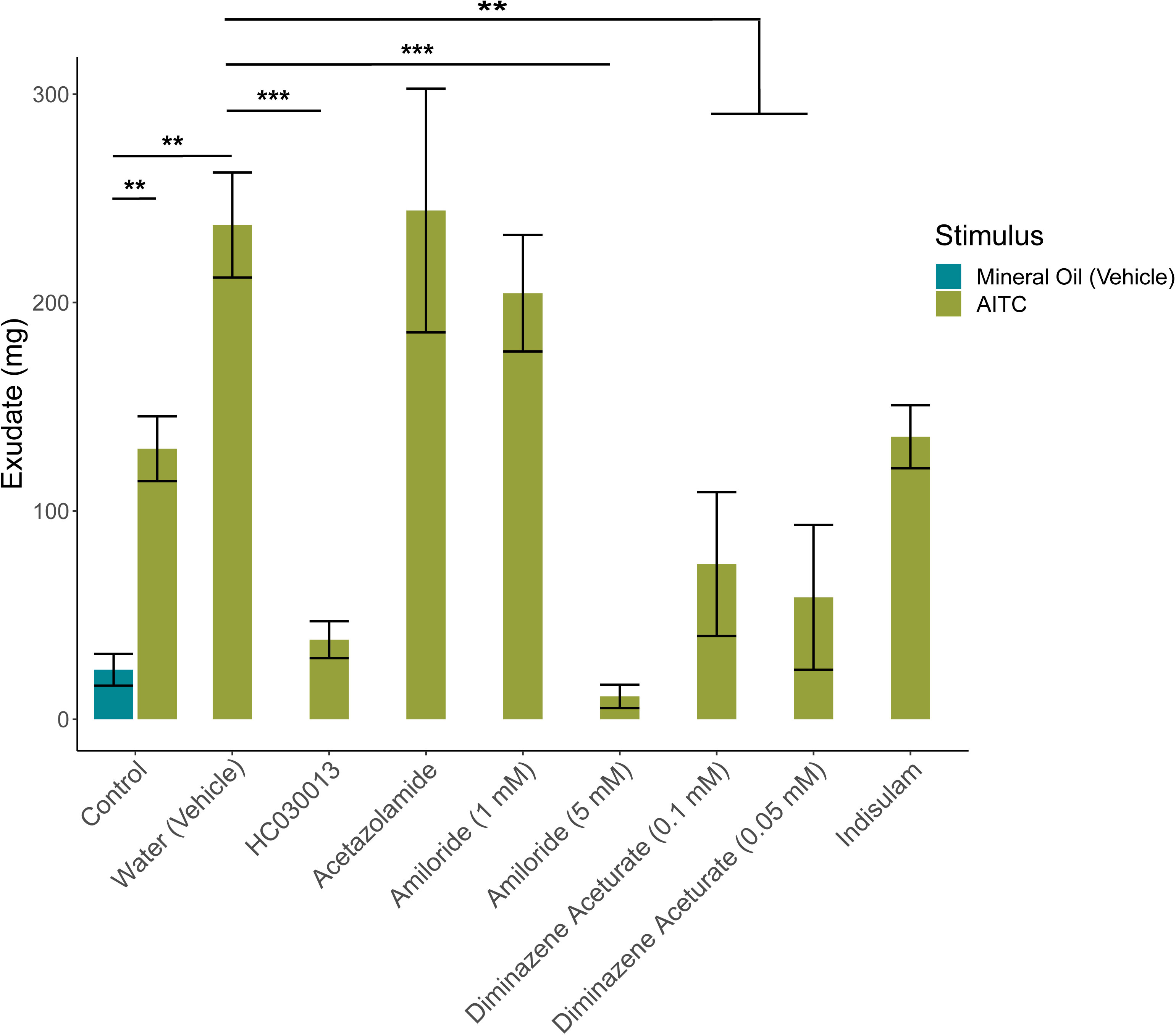
Figure 8 AITC induced exudate excretion after treatment with blockers. AITC significantly increased exudate production (p < 0.01, n=8) and was altered by blocker treatment (p < 0.0001, n=4) compared to control (n=4) via two-way ANOVA. The response to AITC was significantly muted by the TRPA1 blocker HC030013 (p<0.0001, n=7), 5mM amiloride (p<0.0001, n=4), 0.05mM diminazene aceturate (p<0.0001, n=5) and 0.1mM diminazene aceturate (p<001, n=5) via Tukey’s HSD test; while acetazolamide (n=4) and indisulam (n=5) did not significantly alter exudate production. Graphed values are means ± SEM; * p <0.05, ** p <0.01, *** p <0.001.
Organic acids
Carbonic acid is a weak acid; like carbonic acid, other weak acids are also known to stimulate nociceptors in animals (Wang et al., 2011). We decided to test whether earthworms were also sensitive to a series of other weak acids. Indeed, they excreted significantly more exudate in response to increasing concentrations of formic acid, acetic acid, and propionic acid (p<0.0001, n=3-8, two-way ANOVA) (Figure 9A). This response was significantly muted in a dosage-dependent manner after treatment with the sodium channel and ASIC blocker amiloride (p<0.0001, n=4-8, two-way ANOVA) (Figure 9B). This result supports the hypothesis that the detection of other weak acids also requires channels from the ENaC/DEG family.
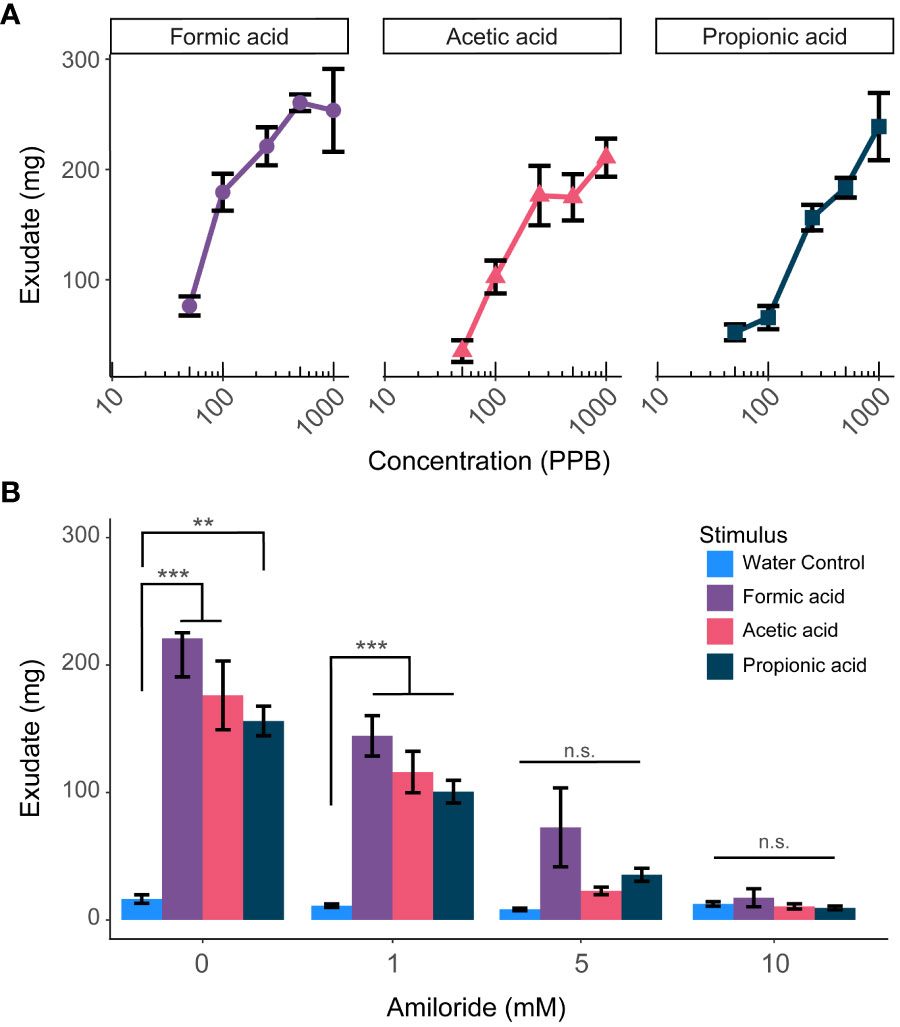
Figure 9 Earthworms produce exudate in response to weak organic acids. (A) Earthworms excrete significantly more exudate in response to increasing concentrations of formic acid, acetic acid, and propionic acid (p<0.0001, two-way ANOVA). (B) This response is significantly muted by amiloride treatment in a dosage-dependent manner (p<0.0001, two-way ANOVA). Graphed values are means ± SEM (n=4-8); * p <0.05, ** p <0.01, *** p <0.001 via Tukey’s HSD test. n.s., not significant.
Discussion
This study demonstrates high CO2 tolerance in D. veneta and provides initial evidence of the molecular mechanisms behind CO2 detection in this species, giving insight into how earthworms detect chemicals in their environment and contributing to ongoing investigations into mechanisms of CO2 detection across species. The chemosensory capabilities of earthworms were famously discussed by Darwin and have been the subject of anatomical, physiology and most often ecological studies since that time. There is, however, a comparative dearth of publications that examine earthworm biology with an eye towards identifying the molecular mechanisms that mediate their physiology and behavior (Stürzenbaum et al., 2009). In fact, we believe that this study is the first to use modern sequencing technologies to elucidate the molecular mechanism responsible for any type of earthworm chemosensation.
CO2 aversion and tolerance
Because earthworms excrete exudate in response to noxious stimuli (Heredia et al., 2008; Heredia Rivera et al., 2020), we used the amount of exudate produced as a proxy for aversion in the exudate assay. We found that CO2 is noxious to earthworms in a dosage-dependent manner (Figure 1C). Many species find CO2 noxious at high concentrations, including rats, nematodes, and fruit flies (Hallem and Sternberg, 2008; van Breugel et al., 2018; Améndola and Weary, 2019). High concentrations of CO2 also anesthetize many species, including rats and fruit flies (Seiger and Kink, 1993; Danneman et al., 1997), but we did not observe this in earthworms even with 100% CO2. Low concentrations of CO2 (ex: 25%) produced no significant response in earthworms, though such concentrations are noxious in other species including rats (25%; Améndola and Weary, 2019), nematodes (10%; Hallem and Sternberg, 2008), and fruit flies (5% when not foraging; van Breugel et al., 2018). Concentrations as high as 45% have been used in human irritation studies, suggesting similar tolerance, although in a much larger animal (Hummel et al., 1998; Shusterman and Avila, 2003). Earthworms may be resistant to high CO2 concentrations due to high exposure in their environment, with mechanistic adaptations underlying such resistance.
Increasing CO2 and therefore increasing carbonic acid is usually considered a proxy measure for hypoxia. There is a precedent for subterranean organisms to be more tolerant of CO2 because of their burrows being hypoxic environments. Naked-mole rats have a proton-insensitive ASIC3 channel among other adaptations to their high-CO2 environment (Schuhmacher et al, 2018); earthworms may have similar adaptations. While subterranean CO2 levels are estimated to be lower than what earthworms are responding to in this study (0.04% to 13.0%), those estimates vary widely (Amundson and Davidson, 1990; Scott, 2011). Additionally, earthworms are often observed to aggregate into balls of 6 to 12 inches in diameter containing hundreds of individuals (Gates, 1961). CO2 levels in a subterranean ball of earthworms may exceed the general estimates of subterranean CO2 in soil; we are not aware of any studies that attempt to model or measure CO2 levels found in aggregates of earthworms. However, we do expect a writhing mass of metabolizing animal tissue to constantly consume oxygen and generate CO2 that would not easily diffuse in a subterranean environment. Perhaps together, earthworms produce CO2 levels consistent with those tested here and CO2 might even be an important chemical signal that limits the size of earthworm aggregates.
Earthworms feed on soil, gaining nutrients from decaying organic matter, bacteria, nematodes, and other microfauna (Curry and Schmidt, 2007), many of which emit CO2. Low concentrations of CO2 may also be appetitive rather than aversive; this is true of mosquitoes and, when in a foraging state, fruit flies (van Breugel et al., 2018; Spanoudis et al., 2020). Low concentrations may also use different mechanisms than aversive high concentrations. Insects like mosquitoes and fruit flies, however, detect CO2 through ionotropic gustatory receptors, which we found no evidence of in the D. veneta transcriptome.
CO2 adaptations
Earthworm mucus, which coats the epithelium under basal conditions, keeps the skin moist for respiration, aids with locomotion, and acts as a buffer (Schrader, 1994; Zhang et al., 2016). This mucus consists of mucin proteins secreted by epithelial cells, mainly goblet cells (Cunha et al., 2011; Rubin, 2014). While little is known about the physiology of earthworm goblet cells, they are well-characterized in the vertebrate respiratory epithelium, which earthworm epithelium resembles histologically (Cunha et al., 2011) and on the ultrastructual level (Sundararaman and Gupta, 1992). If the physiology of goblet cells is as highly conserved between the two tissues as the anatomy, then mucin secretion is likely SNARE-complex dependent exocytosis (Adler et al., 2013), with mucus thickness influenced by the ions transported out of epithelial cells, particularly Cl- (Rogers, 2007).
Exudate, which we define here as the substance excreted in response to noxious stimuli beyond the basal mucus coating, acts as an alarm signal to other earthworms and is attractive to some predators, such as garter snakes (Jiang et al., 1990). Protein and carbohydrate-rich mucus vesicles containing auto-fluorescent membrane-bound chloragosome granules are excreted from between the earthworm’s segments and lubricated by coelomic fluid (Roots and Johnston, 1966; Heredia et al., 2008; Zhang et al., 2016; Guhra et al., 2020). Studies in the closely related Eisenia fetida show that the vesicles form strands outside the body, which differs from the globules we observed in D. veneta (Heredia et al., 2008). Exudate chemical composition also differs between earthworm species, including in D. veneta α-nicotinamide riboside and the organic acids fumarate, succinate, malate, and α-ketoglutarate (Bundy et al., 2001; Rochfort et al., 2017).
It is unclear to what extent mucus and exudate are the same. Many studies electrically or chemically stimulate earthworms to release exudate in order to collect and study mucus (Jiang et al., 1990; Zhang et al., 2016), and E. fetida excretes small amounts of exudate proteins under basal conditions (Heredia Rivera et al., 2020; Heredia et al., 2008). The most recent research in the field has shown that exudate produced in response to varying intensities of electrical stimuli differ in pH and concentration of amino acids, nitrogen, potassium, and phosphorus (Huan et al., 2023). Our transcriptome data suggest that homologs to secreted CAVI may be one component of both exudate and mucus (Figure 2, S1, transcript ID: 67832_c1_g1_i5 and 76341_c1_g1_i1, 76341_c1_g1_i17). CAVI or gustin is secreted in saliva and airway surface liquid in mammals and is known to play a role in chemosensory perception (Leinonen et al., 2004; Fábián et al., 2015).
In our study, earthworms rinsed with deionized water produced more exudate in response to CO2 compared to untreated worms (Figure 1C). We hypothesize that the water washed off the earthworms’ mucus. A scant number of studies have thoroughly examined or compared the composition and the chemical properties of earthworm mucus and exudate. We predict, however, that mucus may have neutralized the low-pH environment created from CO2 and water by carbonic anhydrase. Rinsing the mucus away with deionized water likely allowed more CO2 to dissolve and be converted to bicarbonate and protons by carbonic anhydrase, producing more protons to activate receptors. Alternatively, mucus may have simply physically blocked the epithelium from irritants. That treatment with water also increased the aversive response to AITC supports this hypothesis but does not preclude both mechanisms from contributing to the observed response. These competing hypotheses emphasize the need for more research focusing on the molecular mechanisms that underlie earthworm chemical senses.
Earthworm carbonic anhydrases
While we acknowledge that these concentrations of CO2 are much higher than what an earthworm is likely to encounter in its subterranean environment (up to 13% CO2) (Amundson and Davidson, 1990), the “non-ecological” concentrations used in this study allowed for investigation of the mechanisms behind CO2 aversion. Repetition of the exudate assay after exposure to blockers demonstrated that carbonic anhydrase is necessary for aversion to high concentrations of CO2, as the general carbonic anhydrase inhibitor acetazolamide and the carbonic anhydrase IX/XII inhibitor indisulam both significantly muted responses to CO2 (Figure 7A). Earthworms thus likely have functioning carbonic anhydrases and may have CAIX and CAXII orthologues. That the CAIX/XII inhibitor U-104 and CAIX inhibitor S4 had little effect on CO2-induced irritation, however, may contradict this theory or suggest that the CAIX/XII orthologues have diverged such that these blockers are no longer effective at inhibiting the enzyme or that the isoforms more closely resemble CAIX.
In mammals, CAXII, CAII, CAVB, and CAIV are found in the human nasal mucosa (Tarun et al., 2003), and CAIV is required for sour taste responses to carbon dioxide (Chandrashekar et al., 2009). The CAII/IV inhibitor topiramate had no significant effect in our study, suggesting that CAII and CAIV orthologues are either not involved in earthworm CO2 detection, or are so divergent from those of organisms on which topiramate has been tested that the blocker simply does not have an effect. CAVI, one of the only known secreted isoforms, is found in mammalian saliva and is associated with taste (Henkin et al., 1999; Fábián et al., 2015). We notably found CAVI orthologues in the D. veneta epithelial transcriptome but did not have access to a CAVI inhibitor for pharmacological studies. Since CAVI is secreted protein, it is likely that rinsing the worms with water removed any CAVI present in the epithelial surface, raising the possibility that it might be responsible for the differential response seen before and after rising (Figure 1C). It is nonetheless likely that earthworms secrete a CAVI orthologue in their mucus or exudate that plays some role in CO2 or acid detection.
Multiple carbonic anhydrase isoforms may contribute to earthworms’ response to CO2; this would explain the larger effect by the general carbonic anhydrase inhibitor acetazolamide compared to the inhibitors of single isoforms. Additionally, given the relatively low percentage identity of earthworm CA to its homologs, that acetazolamide retained its effectiveness in muting the earthworms response to CO2 validates it at a broad-spectrum CA inhibitor. The biological redundancy observed in earthworms response to CO2 is likely indicative of the general importance of CO2 detection to all animals, or could be evidence that carbonic anhydrases are of particular importance to the survivability of earthworms. That carbonic anhydrase is required for a response to CO2 also suggests that earthworms’ sensory receptors detect protons or bicarbonate rather than CO2.
Mechanisms of CO2 detection
Earthworm sensory cells are found on their epithelium, either alone as solitary chemoreceptor cells (SCCs) or grouped into epithelial sensory organs (ESOs), which all project to a ventral nerve cord (Hess, 1925; Csoknya et al., 2005; Kiszler et al., 2012). Receptors activated by CO2, bicarbonate, or protons are likely found on the membranes of these cells. There is evidence of epithelial sodium channels including acid-sensing ion channels (Figure 4), receptor-type guanylate cyclases (Figure 3), otopetrins (Figure 5), and TRPA1s (Figure 6) in the D. veneta transcriptome. Guanylate cyclases, OTOP channels, and TRPA1 channels are likely not required for CO2 detection in D. veneta, as their blockers did not significantly affect earthworms’ responses to CO2 (Figure 7B). Because carbonic anhydrase is required for CO2 aversion (Figure 7A) and there were no matches for the gustatory receptor family that detects CO2 in arthropods, it is also unlikely that CO2-specific sensory cells are required.
Amiloride, however, did mute the exudate response to CO2, suggesting that an epithelial sodium channel (ENaC) is required for CO2 detection and aversion (Figure 7B). The ASIC3 blocker diminazene aceturate also muted responses to CO2, but the role of ASIC3 cannot be confirmed because it also muted the response to the positive control AITC, suggesting that it may impact the mechanism of exudate excretion rather than CO2 detection (Figure 8). High concentrations of amiloride also muted responses to AITC, but, because the ENaC blocker amiloride is also a TRPA1 blocker at high concentrations (Banke, 2011), this response was somewhat expected. Low concentrations of amiloride muted responses to CO2 but not AITC, further suggesting the role of the ENaC superfamily in CO2 detection.
This is corroborated by the responses to organic acids, which were also muted by amiloride (Figure 8), suggesting that the same mechanism is responsible for acid detection and CO2 detection. Formic acid, acetic acid, and propionic acid are all weak acids like carbonic acid. That amiloride muted the response to these three organic acids as well as to CO2 provides additional evidence that the mechanism of CO2 detection in D. veneta detects a product of the carbonic anhydrase reaction (carbonic acid, protons, or bicarbonate; Figure 1A) and that an ASIC homolog is required for this response.
Future research and conclusions
Earthworms are critical to many terrestrial ecosystems, yet we know little about how they make decisions about chemicals in their environment. Additional genomic research in D. veneta and related earthworm species is a necessary next step to understanding earthworm chemical ecology, and high throughput sequencing of D. veneta RNA reported here provides the opportunity to do so. RNAi or CRISPR studies are the logical next step to corroborate the pharmacological evidence for the roles of carbonic anhydrase and ENaCs in earthworm CO2 detection presented in this study. These methods, however, are entirely dependent on discovering a reliable technique for collecting very early-stage earthworm embryos, and we have yet to identify any protocols to do so in the literature—another datum highlighting the need to focus more basic cellular biology research on these organisms. Phylogenetic analyses of chemosensory genes across annelids will continue to provide insight into patterns of chemosensory evolution, potentially revealing adaptations to earthworms’ low-light, high-CO2 environment.
CO2 is a critical signaling molecule that earthworms encounter at high concentrations. This research could thus contribute to a necessary understanding of what attracts and repels earthworms, which has potential applications in agriculture and invasive species management. Earthworms have critical roles in soil fertility, and this research may consequently have additional implications in ecosystem health and biodiversity. Research on responses to CO2 may be of particular importance in the context of climate change: as CO2 concentrations in the atmosphere increase, CO2 concentrations in the soil may increase as well. These results suggest that increasing concentrations of CO2 in the soil won’t be aversive to earthworms, demonstrating their potential resilience to a changing environment. Earthworm biodiversity is influenced by elevation and climate (Phillips et al., 2019). Future studies may also investigate how variations in soil CO2 impact biodiversity; earthworm avoidance and attraction to soil CO2 in nature may differ between species and impact ecosystems in ways not detectable by this study. This research also has potential applications in the assessment and treatment of acidic soil, as our data supports previous studies describing earthworm mucus as an effective buffer (Schrader, 1994; Zhang et al., 2016).
These data may also provide insight into CO2 detection and chemosensation across species. Because earthworms respire (e.g. “breathe”) through their skin, their entire body is a respiratory epithelium (Karaca, 2011). This makes them an excellent model for chemosensation in a respiratory epithelium across invertebrate and vertebrate species, including CO2 and acid detection. Our investigation of this topic has contributed to our understanding of the mechanisms earthworms use to detect carbon dioxide and provides yet another example of how these molecular mechanisms are highly conserved among metazoa.
Data availability statement
The RNA-sequencing datasets generated for this study can be found in the NIH's National Library of Medicine's National Center for Biotechnology Information's Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), Bioproject ID PRJNA1026068. The raw exudate data and scripts used to analyze both the RNA-sequencing and exudate assay data can be found in the following GitHub repository: https://github.com/JakeSaunders/Smith_etal_2023_Earthworm_CO2.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
ES, KA, EJ, WS, and CS contributed to the conception and design of the study. ES developed the exudate assay and with JR, SL, and DH, conducted experiments with that assay. KA and CS optimized the RNA extraction protocol. KA, SL, and CS organized and analyzed transcriptomic data. ES wrote the first draft of the manuscript with major input from CJS. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by NSF IOS1355097 to EJ, and grants from the Wake Forest University Center for Molecular Signaling to EJ, WS, and CS. Additional funds were provided by the Wake Forest University Provost’s Office and Wake Forest University’s Department of Biology.
Acknowledgments
Computations for this study were performed using the Wake Forest University (WFU) High-Performance Computing Facility, a centrally managed computational resource available to WFU researchers including faculty, staff, students, and collaborators (Information Systems and Wake Forest University, 2021). Some data and text included in this manuscript originally appeared in ES’s master’s thesis (Smith, 2021).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2023.1202410/full#supplementary-material
References
Adler K., Tuvim M., Dickey B. (2013). Regulated mucin secretion from airway epithelial cells. Front. Endocrinol. 4. doi: 10.3389/fendo.2013.00129
Ai M., Blais S., Park J.-Y., Min S., Neubert T. A., Suh G. S. B. (2013). Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila. J. Neurosci. 33 (26), 10741–10749. doi: 10.1523/Jneurosci.5419-12.2013
Ai M., Min S., Grosjean Y., Leblanc C., Bell R., Benton R., et al. (2010). Acid sensing by the Drosophila olfactory system. Nature 468 (7324), 691–695. doi: 10.1038/nature09537
Agnihotri A. R., Roy A. A., Joshi R. S. (2016). Gustatory receptors in Lepidoptera: chemosensation and beyond. Insect Molecular Biology 25, 519–529. doi: 10.1111/imb.12246
Akiba Y., Mizumori M., Kuo M., Ham M., Guth P. H., Engel E., et al. (2008). CO2 chemosensing in rat oesophagus. Gut 57, 1654–1664. doi: 10.1136/gut.2007.144378
Améndola L., Weary D. M. (2019). Evidence for consistent individual differences in rat sensitivity to carbon dioxide. PloS One 14, e0215808. doi: 10.1371/journal.pone.0215808
Amundson R. G., Davidson E. A. (1990). Carbon dioxide and nitrogenous gases in the soil atmosphere. J. Geochem. Exploration 38, 13–41. doi: 10.1016/0375-6742(90)90091-N
Bandell M., Story G. M., Hwang S. W., Viswanath V., Eid S. R., Petrus M. J., et al. (2004). Noxious cold ion channel TRPA1 is activated by pungent compounds and bradykinin. Neuron 41, 849–857. doi: 10.1016/s0896-6273(04)00150-3
Banerjee S., Deshpande P. A. (2016). On origin and evolution of carbonic anhydrase isozymes: A phylogenetic analysis from whole-enzyme to active site. Comput. Biol. Chem. 61, 121–129. doi: 10.1016/j.compbiolchem.2016.01.003
Banke T. G. (2011). The dilated TRPA1 channel pore state is blocked by amiloride and analogues. Brain Res. 1381, 21–30. doi: 10.1016/j.brainres.2011.01.021
Bautista D. M., Jordt S.-E., Nikai T., Tsuruda P. R., Read A. J., Poblete J., et al. (2006). TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell 124, 1269–1282. doi: 10.1016/j.cell.2006.02.023
Bodenhofer U., Bonatesta E., Horejs-Kainrath C., Hochreiter S. (2015). msa: an R package for multiple sequence alignment. Bioinformatics 31, 3997–3999. doi: 10.1093/bioinformatics/btv494
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics, 30(15):2114–20. doi: 10.1093/bioinformatics/btu170
Bryant D. M., Johnson K., DiTommaso T., Tickle T., Couger M. B., Payzin-Dogru D., et al. (2017). A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 18 (3), 762–776. doi: 10.1016/j.celrep.2016.12.063
Bundy J. G., Osborn D., Weeks J. M., Lindon J. C., Nicholson J. K. (2001). An NMR-based metabonomic approach to the investigation of coelomic fluid biochemistry in earthworms under toxic stress. FEBS Lett. 500, 31–35. doi: 10.1016/S0014-5793(01)02582-0
Carattino M. D., Montalbetti N. (2020). Acid-sensing ion channels in sensory signaling. Am. J. Physiol. Renal Physiol. 318, F531–F543. doi: 10.1152/ajprenal.00546.2019
Chandrashekar J., Yarmolinsky D., von Buchholtz L., Oka Y., Sly W., Ryba N. J. P., et al. (2009). The taste of carbonation. Science 326, 443–445. doi: 10.1126/science.1174601
Charif D., Lobry J. (2007). “SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis,” in Structural approaches to sequence evolution: Molecules, networks, populations, series Biological and Medical Physics, Biomedical Engineering. Eds. Bastolla U., Porto M., Roman H., Vendruscolo M. (New York: Springer Verlag), 207–232.
Chu X., Kc P., Ian E., Kvello P., Liu Y., Wang G. R., et al. (2020). Neuronal architecture of the second-order CO2 pathway in the brain of a noctuid moth. Sci. Rep. 10, 19838. doi: 10.1038/s41598-020-76918-1
Csoknya M., Takács B., Koza A., Dénes V., Wilhelm M., Hiripi L., et al. (2005). Neurochemical characterization of nervous elements innervating the body wall of earthworms (Lumbricus, Eisenia): immunohistochemical and pharmacological studies. Cell Tissue Res. 321, 479–490. doi: 10.1007/s00441-005-1134-4
Cummins E. P., Strowitzki M. J., Taylor C. T. (2020). Mechanisms and consequences of oxygen and carbon dioxide sensing in mammals. Physiol. Rev. 100, 463–488. doi: 10.1152/physrev.00003.2019
Cunha L., Campos I., Montiel R., Rodrigues A., Morgan A. J. (2011). Morphometry of the epidermis of an invasive megascoelecid earthworm (Amynthas gracilis, Kinberg 1867) inhabiting actively volcanic soils in the Azores archipelago. Ecotoxicol. Environ. Saf. 74, 25–32. doi: 10.1016/j.ecoenv.2010.08.004
Curry J. P., Schmidt O. (2007). The feeding ecology of earthworms – A review. Pedobiologia 50, 463–477. doi: 10.1016/j.pedobi.2006.09.001
Danneman P. J., Stein S., Walshaw S. O. (1997). Humane and practical implications of using carbon dioxide mixed with oxygen for anesthesia or euthanasia of rats. Lab. Anim. Sci. 47, 376–385.
Detweiler N. D., Vigil K. G., Resta T. C., Walker B. R., Jernigan N. L. (2018). Role of acid-sensing ion channels in hypoxia- and hypercapnia-induced ventilatory responses. PloS One 13, e0192724. doi: 10.1371/journal.pone.0192724
Edwards C. (2004). “Chapter 1, the importance of earthworms as key representatives of the soil fauna,” in Earthworm Ecology, 2nd (Danvers, MA: CRC Press LLC), 3–9.
Eid S. R., Crown E. D., Moore E. L., Liang H. A., Choong K.-C., Dima S., et al. (2008). HC-030031, a TRPA1 selective antagonist, attenuates inflammatory- and neuropathy-induced mechanical hypersensitivity. Mol. Pain 4, 48. doi: 10.1186/1744-8069-4-48
Fábián T., Beck A., Fejérdy P., Hermann P., Fábián G. (2015). Molecular mechanisms of taste recognition: considerations about the role of saliva. Int. J. Mol. Sci. 16 (12), 5945–5974. doi: 10.3390/ijms16035945
Fang X., Seim I., Huang Z., Gerashchenko M. V., Xiong Z., Turanov A. A., et al. (2014). Adaptations to a subterranean environment and longevity revealed by the analysis of mole rat genomes. Cell Rep. 8, 1354–1364. doi: 10.1016/j.celrep.2014.07.030
Gates G. E. (1961). Ecology of some earthworms with special reference to seasonal activity. Am. Midland Naturalist 66 (1), 61. doi: 10.2307/2422868
Guhra T., Stolze K., Schweizer S., Totsche K. U. (2020). Earthworm mucus contributes to the formation of organo-mineral associations in soil. Soil Biol. Biochem. 145, 107785. doi: 10.1016/j.soilbio.2020.107785
Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hallem E. A., Spencer W. C., McWhirter R. D., Zeller G., Henz S. R., Rätsch G., et al. (2011). Receptor-type guanylate cyclase is required for carbon dioxide sensation by Caenorhabditis elegans. Proc. Natl. Acad. Sci. United States America 108, 254–259. doi: 10.1073/pnas.1017354108
Hallem E. A., Sternberg P. W. (2008). Acute carbon dioxide avoidance in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 105, 8038–8043. doi: 10.1073/pnas.0707469105
Hanukoglu I., Hanukoglu A. (2016). Epithelial sodium channel (ENaC) family: Phylogeny, structure-function, tissue distribution, and associated inherited diseases. Gene 579, 95–132. doi: 10.1016/j.gene.2015.12.061
Henkin R., Brian M., Raghunath A. (1999). Decreased parotid saliva gustin/carbonic anhydrase VI secretion: an enzyme disorder manifested by gustatory and olfactory dysfunction. Am. J. Med. Sci. 318 (6), 380–391. doi: 10.1016/S0002-9629(15)40663-9
Henry R. P. (1996). Multiple roles of carbonic anhydrase in cellular transport and metabolism. Annu. Rev. Physiol. 58, 523–538. doi: 10.1146/annurev.ph.58.030196.002515
Heredia R. B., Dueñas S., Castillo L., Ventura J. J., Silva Briano M., Posadas del Rio F., et al. (2008). Autofluorescence as a tool to study mucus secretion in Eisenia foetida. Comp. Biochem. Physiol. Part A.: Mol. Integr. Physiol. Fifth Special Issue CBP Dedicated “The Face Latin Am. Comp. Biochem. Physiol”. 151, 407–414. doi: 10.1016/j.cbpa.2007.01.726
Heredia Rivera B., Rodríguez M. G., Rodríguez-Heredia M., et al. (2020). Characterisation by Excitation-emission matrix fluorescence spectroscopy of pigments in mucus secreted of earthworm eisenia foetida exposed to lead. J Fluoresc. 30, 725–733. doi: 10.1007/s10895-020-02533-y
Hess W. N. (1925). Nervous system of the earthworm, lumbricus terrestris L. J. Morphol. 40, 235–259. doi: 10.1002/jmor.1050400203
Huan H., Wang X., Chu Z., Yu X., Fan T., Li G., et al. (2023). Compositional changes and ecological characteristics of earthworm mucus under different electrical stimuli. Sci. Rep. 13, 2332. doi: 10.1038/s41598-023-29125-7
Hummel T., Kraetsch H. G., Pauli E., Kobal G. (1998). Responses to nasal irritation obtained from the human nasal mucosa. Rhinology 36, 168–172.
Information Systems and Wake Forest University (2021). WFU high performance computing facility. doi: 10.57682/g13z-2362
Jiang X. C., Inouchi J., Wang D., Halpern M. (1990). Purification and characterization of a chemoattractant from electric shock-induced earthworm secretion, its receptor binding, and signal transduction through the vomeronasal system of garter snakes. J. Biol. Chem. 265, 8736–8744. doi: 10.1016/S0021-9258(19)38950-1
Karaca A. (2011). Biology of earthworms, soil biology. Springer Berlin Heidelberg. pp, 141–158. doi: 10.1007/978-3-642-14636-7_9
Kellenberger S., Schild L. (2002). Epithelial sodium channel/degenerin family of ion channels: A variety of functions for a shared structure. Physiol. Rev. 82, 735–767. doi: 10.1152/physrev.00007.2002
Kiszler G., Varhalmi E., Berta G., Molnar L. (2012). Organization of the sensory system of the earthworm Lumbricus terrestris (Annelida, Clitellata) visualized by DiI. J. Morphol. 273, 737–745. doi: 10.1002/jmor.20018
Kuhn M. (2016). Molecular physiology of membrane guanylyl cyclase receptors. Physiol. Rev. 96, 751–804. doi: 10.1152/physrev.00022.2015
Kwon J. Y., Dahanukar A., Weiss L. A., Carlson J. R. (2007). The molecular basis of CO2 reception in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 104 (9), 3574–3578. doi: 10.1073/pnas.0700079104
Leinonen J. S., Saari K. A., Seppänen J. M., Myllylä H. M., Rajaniemi H. J. (2004). Immunohistochemical demonstration of carbonic anhydrase isoenzyme VI (CA VI) expression in rat lower airways and lung. J. Histochem. Cytochem. 52 (8), 1107–1112. doi: 10.1369/jhc.4a6282.2004
Li W.-G., Xu T.-L. (2011). ASIC3 channels in multimodal sensory perception. ACS Chem. Neurosci. 2, 26–37. doi: 10.1021/cn100094b
Lindskog S. (1997). Structure and mechanism of carbonic anhydrase. Pharmacol. Ther. 74, 1–20. doi: 10.1016/S0163-7258(96)00198-2
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Makino C. L., Duda T., Pertzev A., Isayama T., Geva P., Sandberg M. A., et al. (2019). Modes of accessing bicarbonate for the regulation of membrane guanylate cyclase (ROS-GC) in retinal rods and cones. eNeuro 6, 0393–18. doi: 10.1523/ENEURO.0393-18.2019
McKenzie S. K., Fetter-Pruneda I., Ruta V., Kronauer D. J. C. (2016). Transcriptomics and neuroanatomy of the clonal raider ant implicate an expanded clade of odorantreceptors in chemical communication. Proc. Natl. Acad. Sci. U.S.A. 113, 14091–14096. doi: 10.1073/pnas.1610800113
McKenzie S. K., Kronauer D. J. C. (2018). The genomic architecture and molecular evolution of ant odorant receptors. Genome Res. 28, 1–9. doi: 10.1101/gr.237123.118
Ning C., Yang K., Xu M., Huang L.-Q., Wang C.-Z. (2016). Functional validation of the carbon dioxide receptor in labial palps of Helicoverpa armigera moths. Insect Biochem. Mol. Biol. 73, 12–19. doi: 10.1016/j.ibmb.2016.04.002
Osmakov D. I., Andreev Y. A., Kozlov S. A. (2014). Acid-sensing ion channels and their modulators. Biochem. Moscow. 79, 1528–1545. doi: 10.1134/S0006297914130069
Phillips H. R. P., Guerra C. A., Bartz M. L. C., Briones M. J. I., Brown G., Crowther T. W., et al. (2019). Global distribution of earthworm diversity. Science 366, 480–485. doi: 10.1126/science.aax4851
R Core Team (2021). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Reed E. M., O’Connor M. O., Johnson I. C., Silver W. L., Saunders C. J. (2021). Dendrobaena veneta avoids ethyl pentanoate and ethyl hexanoate, two compounds produced by the soil fungus Geotrichum candidum. PeerJ 9, e12148. doi: 10.7717/peerj.12148
Rhoades J. L., Nelson J. C., Nwabudike I., Yu S. K., McLachlan I. G., Madan G. K., et al. (2019). ASICs mediate food responses in an enteric serotonergic neuron that controls foraging behaviors. Cell 176, 85–97.e14. doi: 10.1016/j.cell.2018.11.023
Rochfort S., Wyatt M. A., Liebeke M., Southam A. D., Viant M. R., Bundy J. G. (2017). Aromatic metabolites from the coelomic fluid of Eisenia earthworm species. Eur. J. Soil Biol. 78, 17–19. doi: 10.1016/j.ejsobi.2016.11.008
Rogers D. F. (2007). Physiology of airway mucus secretion and pathophysiology of hypersecretion. Respir. Care 52, 1134–46.
Roots B. I., Johnston P. V. (1966). The lipids and pigments of the chloragosomes of the earthworm Lumbricus terrestris. L. Comp. Biochem. Physiol. 17, 285–288. doi: 10.1016/0010-406x(66)90027-2
Rubin B. K. (2014). Secretion properties, clearance, and therapy in airway disease. Transl. Respir. Med. 2:6. doi: 10.1186/2213-0802-2-6
Scott K. (2011). Out of thin air: Sensory detection of oxygen and carbon dioxide. Neuron 69, 194–202. doi: 10.1016/j.neuron.2010.12.018
Schrader S. (1994). Influence of earthworms on the pH conditions of their environment by cutaneous mucus secretion. Zoologischer Anzeiger 233 (5-6), 211–219.
Schuhmacher L. N., Callejo G., Srivats S., Smith E. S. J. (2018). Naked mole-rat acid-sensing ion channel 3 forms nonfunctional homomers, but functional heteromers. J. Biol. Chem. 293 (5), 1756–1766. doi: 10.1074/jbc.M117.807859
Seiger M. B., Kink J. F. (1993). The effect of anesthesia on the photoresponses of four sympatric species of Drosophila. Behav. Genet. 23, 99–104. doi: 10.1007/BF01067559
Shams I., Avivi A., Nevo E. (2005). Oxygen and carbon dioxide fluctuations in burrows of subterranean blind mole rats indicate tolerance to hypoxic–hypercapnic stresses. Comp. Biochem. Physiol. Part A.: Mol. Integr. Physiol. 142, 376–382. doi: 10.1016/j.cbpa.2005.09.003
Shusterman D., Avila P. C. (2003). Real-time monitoring of nasal mucosal pH during carbon dioxide stimulation: implications for stimulus dynamics. Chem. Senses 28, 595–601. doi: 10.1093/chemse/bjg050
Silver W. L., Kim A. H., Kim E. Y., Saunders C. J. (2019). A novel T-maze assay to evaluate chemical irritants on Lumbricus terrestris. Appl. Soil Ecol. 133, 186–189. doi: 10.1016/j.apsoil.2018.10.007
Silver W., Kim E., Krivda K., Robertson J., Smith K. (2018). Are TRPA1 channels involved in the detection of chemical irritants by the earthworm, Eisenia hortensis? Chem. Senses 43, 543.
Smith K. (2019). A Molecular Investigation of chemesthesis in Eisenia hortensis through analysis of transient receptor potential channels. [dissertation/master’s thesis] (Winston-Salem (NC: Wake Forest University).
Smith E. J. (2021). Mechanisms of carbon dioxide detection in the earthworm Eisenia hortensis (Winston-Salem (NC: Wake Forest University).
Smith E., Martinez-Velazquez L., Ringstad N. (2013). A chemoreceptor that detects molecular carbon dioxide. J. Biol. Chem. 288, 37071–37081. doi: 10.1074/jbc.M113.517367
Spanoudis C. G., Andreadis S. S., Bray D. P., Savopoulou-Soultani M., Ignell R. (2020). Behavioural response of the house mosquitoes Culex quinquefasciatus and Culex pipiens molestus to avian odours and its reliance on carbon dioxide. Med. Veterinary Entomol. 34, 129–137. doi: 10.1111/mve.12429
Stürzenbaum S. R., Andre J., Kille P., Morgan A. J. (2009). Earthworm genomes, genes and proteins: the (re) discovery of Darwin’s worms. Proc. R. Soc. B.: Biol. Sci. 276 (1658), 789–797. doi: 10.1098/rspb.2008.1510
Suh G. S. B., Wong A. M., Hergarden A. C., Wang J. W., Simon A. F., Benzer S., et al. (2004). A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature 431, 854–859. doi: 10.1038/nature02980
Sun L., Wang H., Hu J., Han J., Matsunami H., Luo M. (2009). Guanylyl cyclase-D in the olfactory CO2 neurons is activated by bicarbonate. PNAS 106, 2041–2046. doi: 10.1073/pnas.0812220106
Sundararaman V., Gupta S. K. (1992). An ultrastructural study of the clitellar epithelium of the earthworm Metaphire posthuma (vail.). Tissue Cell 24 (5), 745–750. doi: 10.1016/0040-8166(92)90046-a
Takahashi N., Mizuno Y., Kozai D., Yamamoto S., Kiyonaka S., Shibata T., et al. (2008). Molecular characterization of TRPA1 channel activation by cysteine-reactive inflammatory mediators. Channels 2, 287–298. doi: 10.4161/chan.2.4.6745
Tarun A. S., Bryant B., Zhai W., Solomon C., Shusterman D. (2003). Gene expression for carbonic anhydrase isoenzymes in human nasal mucosa. Chem. Senses. 28, 621–629. doi: 10.1093/chemse/bjg054
Teng B., Wilson C. E., Tu Y.-H., Joshi N. R., Kinnamon S. C., Liman E. R. (2019). Cellular and neural responses to sour stimuli require the proton channel otop1. Curr. Biol. 29, 3647–3656.e5. doi: 10.1016/j.cub.2019.08.077
Tu Y.-H., Cooper A. J., Teng B., Chang R. B., Artiga D. J., Turner H. N., et al. (2018). An evolutionarily conserved gene family encodes proton-selective ion channels. Science 359, 1047–1050. doi: 10.1126/science.aao3264
van Breugel F., Huda A., Dickinson M. H. (2018). Distinct activity-gated pathways mediate attraction and aversion to CO2 in Drosophila. Nature 564, 420–424. doi: 10.1038/s41586-018-0732-8
Wang Y. Y., Chang R. B., Allgood S. D., Silver W. L., Liman E. R. (2011). A TRPA1-dependent mechanism for the pungent sensation of weak acids. J. Gen. Physiol. 137, 493–505. doi: 10.1085/jgp.201110615
Wang Y. Y., Chang R. B., Liman E. R. (2010). TRPA1 is a component of the nociceptive response to CO2. J. Neurosci. 30, 12958–12963. doi: 10.1523/JNEUROSCI.2715-10.2010
Wickham H. (2016). ggplot2: Elegant Graphics for Data Analysis (Verlag New York: Springer). Available at: https://ggplot2.tidyverse.org.
Wickham H., Averick M., Bryan J., Chang W., McGowan L. D., François R., et al. (2019). Welcome to the tidyverse. J. Open Source Software 4, 1686. doi: 10.21105/joss.01686
Wickham H., François R., Henry L., Müller K., Vaughan D. (2023). dplyr: A Grammar of Data Manipulation. R package version 1.1.4. Available at: https://github.com/tidyverse/dplyr,https://dplyr.tidyverse.org.
Yu G., Smith D., Zhu H., Guan Y., Lam T. T. (2017). ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evolution. 8, 28–36. doi: 10.1111/2041-210X.12628
Keywords: Eisenia hortensis, European nightcrawler, carbonic anhydrases, chemical senses, chemosensory, epithelial sodium channels (ENaCs), degenerin, TRPA1 (transient receptor potential A1)
Citation: Smith EJ, Ryan JL, Lopresti SA, Haghnazari DBS, Anderson KAS, Lipson SJ, Johnson EC, Silver WL and Saunders CJ (2023) Mechanisms of carbon dioxide detection in the earthworm Dendrobaena veneta. Front. Ecol. Evol. 11:1202410. doi: 10.3389/fevo.2023.1202410
Received: 08 April 2023; Accepted: 08 November 2023;
Published: 29 November 2023.
Edited by:
Alberto Maria Cattaneo, Swedish University of Agricultural Sciences, SwedenCopyright © 2023 Smith, Ryan, Lopresti, Haghnazari, Anderson, Lipson, Johnson, Silver and Saunders. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emily Jordan Smith, smithej@bu.edu; emi.jor.smi@gmail.com; Cecil J. Saunders, csaunder@kean.edu; jakesaunders@gmail.com
 Emily Jordan Smith
Emily Jordan Smith Jennifer L. Ryan
Jennifer L. Ryan Sofia A. Lopresti1
Sofia A. Lopresti1  Karleigh A. S. Anderson
Karleigh A. S. Anderson Sarah J. Lipson
Sarah J. Lipson Erik C. Johnson
Erik C. Johnson Wayne L. Silver
Wayne L. Silver Cecil J. Saunders
Cecil J. Saunders